- 1Department of Clinical Laboratory, Jingjiang People’s Hospital, Taizhou, China
- 2Department of General Surgery, Jingjiang People’s Hospital, Taizhou, China
Background: Circular RNAs (circRNAs) have proven as a special subset of endogenous RNAs that are implicated in the tumorigenesis of various cancers. This study sought to evaluate the role of circRNAs in the diagnosis and prognosis of colorectal cancer (CRC).
Methods: The online databases were searched for collecting relevant studies on circRNAs as diagnostic and prognostic biomarkers of CRC. Two researchers independently screened literature, extracted data, and evaluated the bias and risks of included studies. The diagnostic and prognostic indicators were merged and analyzed using STATA 12.0 software, and sources of heterogeneity were traced by the sensitivity analysis and the meta-regression test.
Results: A total of 29 articles representing 2639 CRC patients were included. The pooled sensitivity, specificity, and area under the curve (AUC) of circRNAs in differentiating CRC from non-tumor control were 0.75 (95% CI: 0.69–0.80) and 0.74 (95% CI: 0.69–0.78) and 0.81, respectively. The survival analysis showed that up-regulations of up-regulated circRNAs were significantly related to dismal survival in CRC patients (HR = 2.38, p < 0.001). A stratified analysis showed that the comprehensive diagnostic value of up-regualted circRNAs in CRC was higher than that of down-regualted circRNAs (AUC: 0.83 vs. 0.77; Z test, p < 0.05). The efficacy of tissue-derived circRNAs in the diagnosis of CRC was equal to that of plasma/serum-derived ones (AUC: 0.81 vs. 0.82; Z test, p > 0.05).
Conclusion: Abnormally expressed circRNAs as auxiliary biomarkers present underlying value in the diagnosis and prognosis prediction of CRC.
Introduction
CRC as one of the most common malignant tumors of the digestive tract accounts for 10.2% of total cancer cases—18.1 million new cases worldwide in 2018—according to Global Cancer Statistics 2018 released by WHO, ranking the third (1). As with the 2015 China Cancer report, CRC ranks the fourth and third in morbidity and mortality in the nation, respectively (2). Due to the concealment of early symptoms of the disease, most of the patients initially visit a doctor until the middle and late stage (3). Along with the limited therapeutic effect and multiple relapses, the prognosis of the patients is very poor (4). Therefore, establishing an early diagnostic system for CRC, which means better prognosis and timely adoptions of new and effective therapies, is the preoccupation for reducing the mortality and ameliorating the prognosis. Though endoscopic biopsy combined with histopathology is the gold standard for its diagnosis, such an invasive examination cannot be easily accepted by some patients (5,6). The diagnostic efficacy of traditional biomarkers including carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and CA72-4 are limited (4). A published meta-analysis has shown that the total sensitivity of CA19-9 in the diagnosis of CRC is only from 0.68 to 0.71 (a threshold of 5 μg/L to 10 μg/L), far from the requirements of clinical diagnosis and treatments (7). In this respect, searching for new markers is an important means of early diagnosis of CRC.
CircRNAs are a class of non-coding RNA featuring covalent bindings that form a closed loop with the 3′ and 5′ ends (8,9). CircRNAs, widely expressed in mammalian cells, are tissue-cell specific, structurally stable and sequence-conserved (10). Some types of circRNAs are proven to play roles in the transcription and expression of genes via multiple ways, which play an important role in cell cycle, cell aging and other physiological processes (10,11). Accumulating studies have shown that circRNAs are critical in the occurrence and development of malignant tumors (12,13). At present, many studies have reported that circRNAs have underlying clinical values in the early diagnosis and prognosis evaluation of CRC (14–42). In view of the fact that there are noticeable problems of single-item single-center trials, such as small sample size and large result bias, this study intends to systematically evaluate the potential application value of circRNAs in clinical diagnosis and prognosis evaluation of CRC through a PRISMA-compliant pair-wise meta-analysis.
Materials and Methods
Data Search Strategy
Retrieval data were extracted from papers published in English on PubMed, EMBASE, Web of Science and CNKI databases due November 1st, 2021. The key words encompassed “colorectal cancer”, “colorectal carcinoma”, “carcinoma of colon”, “colorectal neoplasms”, “circular RNA”, “circRNA”, “hsa circ”, “diagnoses”, “diagnosis”, “sensitivity”, “specificity”, “area under the curve”, “AUC”, “ROC curve”, “prognoses”, “prognosis”, “survival”, “overall survival”, “progression free survival”, “hazard ratio”, “OS”, “PFS” and “HR”. Besides, references attached to the paper were also searched manually to prevent omission of any eligible literature.
Inclusion and Exclusion Criteria
Inclusion criteria were listed as follows: 1) case-control studies; 2) studies on the evaluation of diagnostic value and/or prognosis of circRNA in CRC; 3) data of true positive number (TP), false positive number (FP), false negative number (FN) and true negative number (TN) that could be obtained directly or indirectly to construct a 2 × 2 four-grid table for the diagnostic meta-analysis; 4) the prognostic observation indice was total survival time (overall survival, OS), or progression free survival (PFS), for outputting the hazard ratio (HR) and 95% confidence interval (CI) directly or indirectly. Exclusion criteria were defined as follows: 1) a study size of less than 20 cases; 2) data used for statistical analysis were insufficient or unavailable even after contacting original authors; 3) non-English language articles or low quality research.
Data Extraction
All relevant literature was screened by two trained authors. The basic information was extracted independently: name of the first author, date of publication, study population, sample size, the control type, circRNA signatures, detection methods, reference genes, cut-off value settings, follow-up time, as well as values of sensitivity, specificity, AUC, HR, and 95% CI(s) and so forth.
Quality Assessment
The Quality Assessment of Diagnostic Accuracy Studies (QUADAS)-2 tool was adopted to evaluate the quality of references prior to the diagnostic meta-analysis (43). The evaluation system comprised two parts: bias evaluation and applicability. Specifically, the bias evaluation consisted of case selection, index tests, reference standards, and flow and timing, and the applicability evaluation of case selection, trials to be evaluated and gold standard. Each item could be classified as low risk, high risk and unknown, with the corresponding scores of 1, 0, and 0, respectively. The total score of ≥4 (with a full score of 7) indicated that the quality of literature research was high. The case-control study was evaluated according to 8 items of the Newcastle-Ottawa Scale (NOS scale) (44), categorized into study selection, comparability, and outcome. The total rated score of ≥5 (with a full score of 9) suggested that the quality of literature research was high.
Statistical Analysis
Statistical analysis was carried out using MetaDiSc 1.4 and STATA 12.0 software, and the combined-effect indices involved sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), AUC, HR and 95% CIs. The difference in the merged area under the curve (AUC) between groups was compared by Z test. Spearman correlation coefficients were calculated to evaluate the threshold effect using MetaDiSc 1.4, while Cochran’ Q and I2 tests to assess the non-threshold effect using STATA 12.0. The level of significance was set at p < 0.01 or I2 > 50%. When there was no heterogeneity between studies, the statistics could be merged by the fixed-effect model; and when heterogeneity appeared, the statistics would be combined using the random-effect model. The sources of heterogeneity were deeply traced by a sensitivity analysis and a meta-regression test. Publication bias was judged by Deek’s quantitative funnel plot, as well as Begg’s and Egger’s tests, and the significant level was set at p < 0.1. The trim-and-fill method was adopted to assess the possible effect of publication bias (45).
Results
Study Selection and Data Characteristics
Of the initially screened 536 references from the aforesaid online databases according to the retrieval strategy, 29 articles (19 for diagnosis and 14 for prognosis) (14–42) that met the inclusion and exclusion criteria were eventually enrolled for our meta-analysis. The literature retrieval process was depicted in Figure 1.
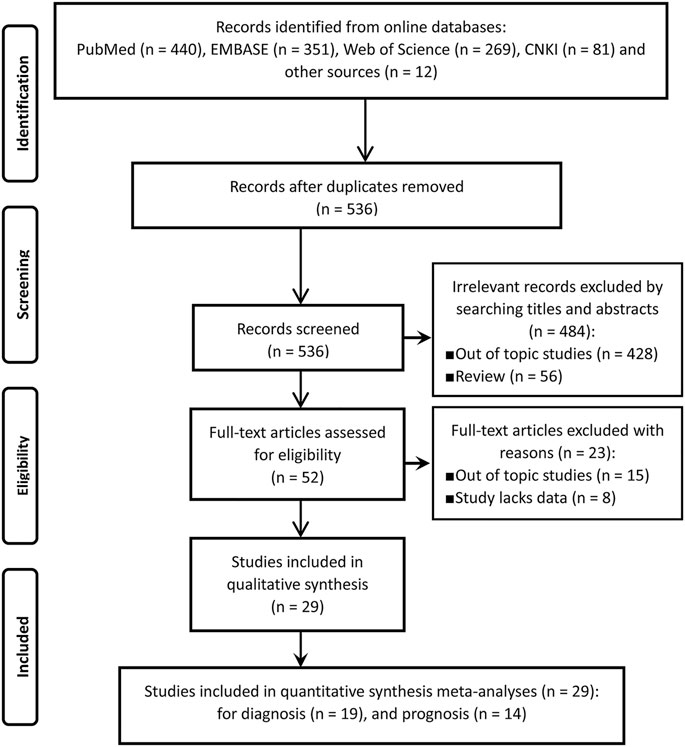
FIGURE 1. The flow chart of literature inclusion and exclusion process according to the PRISMA statement.
The basic characteristics of the 29 articles (14–42) were summarized in Tables 1, 2. A total of 2639 CRC patients and 1471 matched controls were enrolled. The CRC cases were confirmed by pathological examinations and the included control group encompassed healthy subjects and paracancerous controls. All tissue and plasma samples were obtained preoperatively without any other treatment. The included individuals in our study included Asians and Caucasians, and 34 circRNAs were involved in the meta-analysis, of which 25 acting as oncogenes were up-regulated and 9 as tumor-suppressor genes were down-regulated in CRC. The expression levels of circRNAs in CRC were determined by RT-qPCR or RNA sequencing, and GAPDH, 18srRNA, or β-actin mRNAs were used as internal reference genes. Of the 15 included studies on the prognosis of CRC, 9 provided available HRs and 95% CIs, 6 offered relevant data that could indirectly calculate the indices by formulas or prognosis curves. The median follow-up time varied from 1 to 39 months.
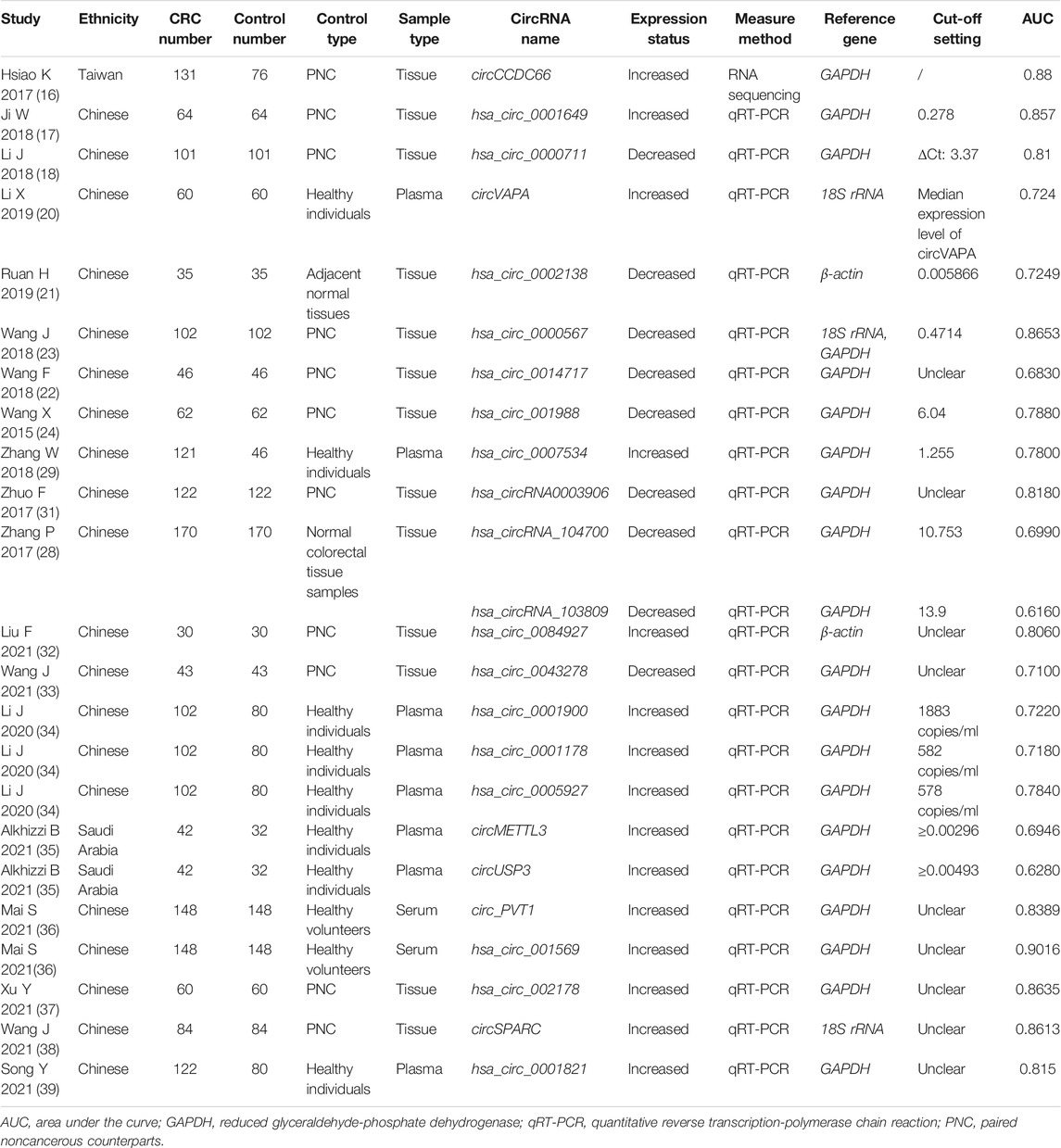
TABLE 1. Basic characteristics of the included studies for assessing diagnostic performances of circRNAs in CRC.
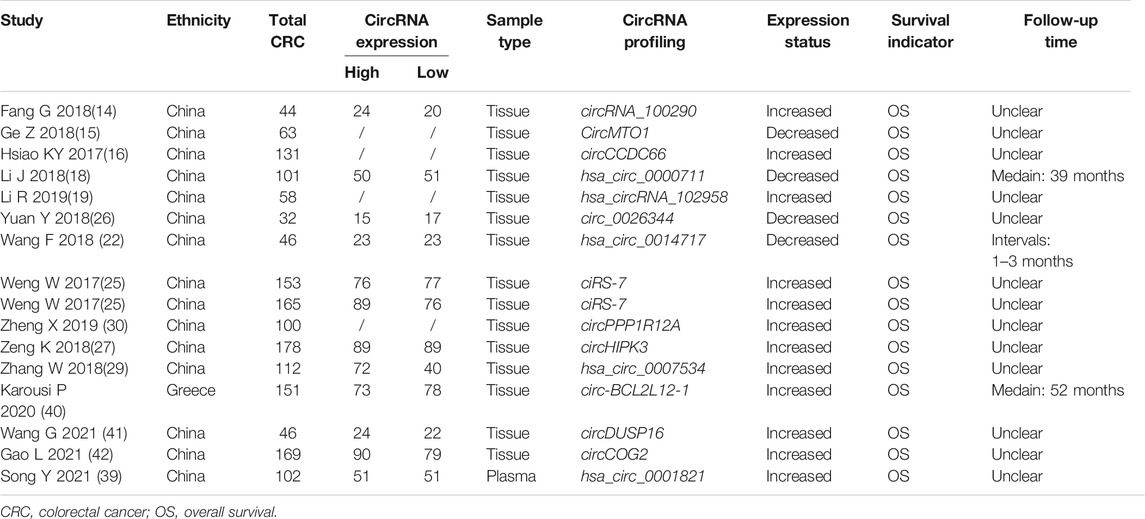
TABLE 2. Basic characteristics of the included studies for the appraisal of prognostic effects of circRNAs on CRC.
Heterogeneity Test
The Spearman correlation coefficient analysis showed a p value of 0.356 (Spearman correlation coefficient: −0.267) in diagnostic meta-analysis, suggesting that there was no heterogeneity resulted from the threshold effect. A p value of <0.001 and I2 of 97.46% for the overall combined diagnostic effect were presented in Cochran’ Q and I2 tests, indicating that substantial heterogeneity existed in the non-threshold effect. No heterogeneity was observed in the combined prognostic meta-analysis (oncogenic circRNAs: I2 = 0.0%, p = 1.000; adjusted effect of the tumor-suppressor circRNAs: I2 = 0.0%, p = 0.994).
Bias Risk Assessments for the Included Studies
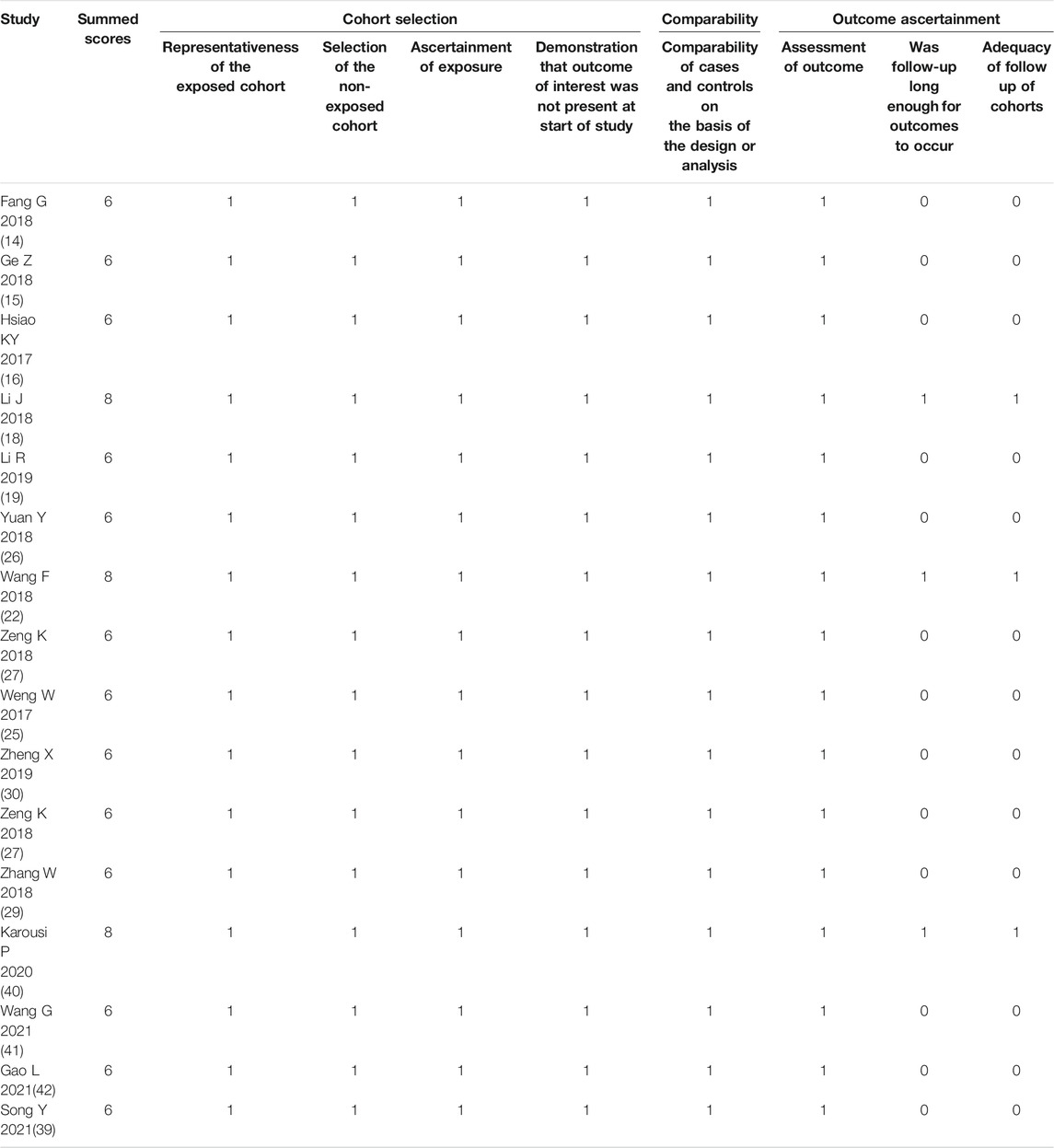
For the diagnostic meta-analysis, the QUADAS-2 scale was used to assess the risk of bias. As a result, the rated scores of all 19 articles were higher than 4 (Table 3), suggesting that the overall quality of the included studies was high. Consistently, the NOS scale showed high scores of over 6 in the observational studies (Table 4), indicating the high quality of the included case-control studies.
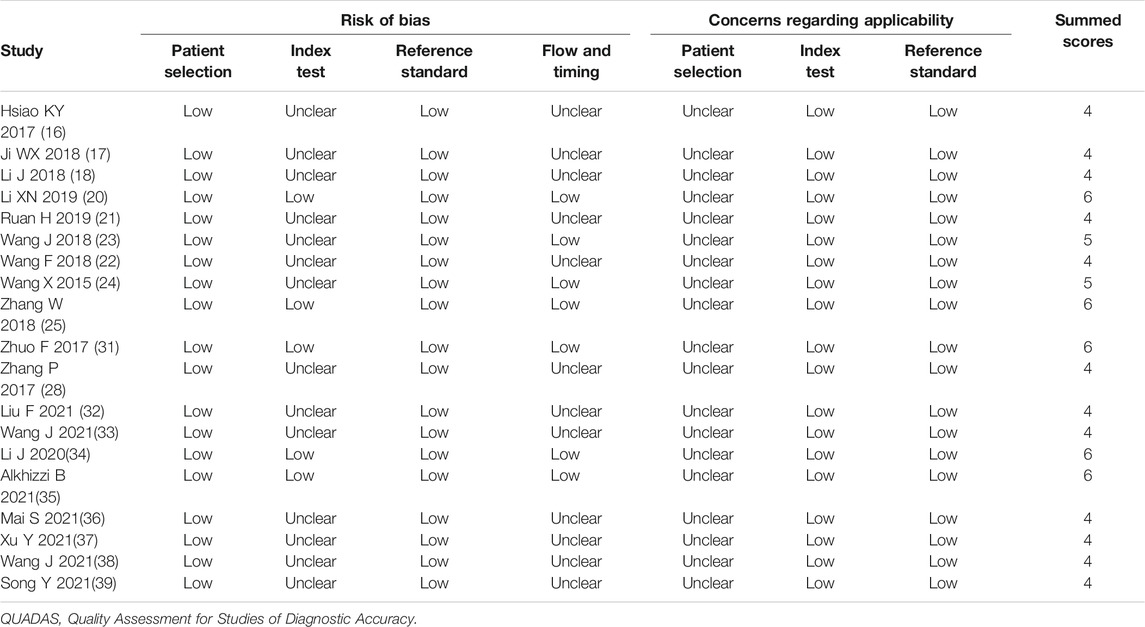
TABLE 3. The study quality and the bias risk in studies on the diagnostic efficacy of circRNAs were assessed by the QUADAS II checklist.
Diagnostic Efficiency
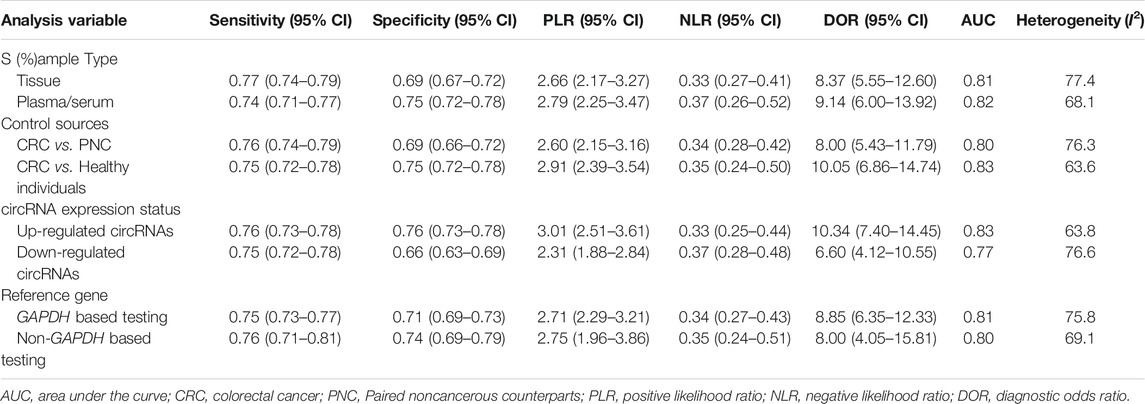
The forest plots showed that the pooled sensitivity, specificity, PLR, NLR, DOR and AUC of circRNAs in the diagnosis of CRC were 0.75 (95% CI: 0.69–0.80), 0.74 (95% CI: 0.69–0.78), 2.87 (95% CI: 2.46–3.34), 0.34 (95% CI: 0.28–0.42), 8.37 (95% CI: 6.32–11.09) and 0.81, as shown in Figure 2. Subgroup analysis showed that the AUC of up-regulated circRNAs in the diagnosis of CRC was higher than that of down-regulated circRNAs (AUC: 0.83 versus 0.77; Z test, p < 0.05), and the former had higher diagnostic specificity and DOR (Table 5). Analysis based on different control sources showed that the efficacy of circRNA expression profile was higher in distinguishing CRC from healthy differentiation than its ability to distinguish CRC from PNC (Table 5). Analysis based on different sample type and reference gene showed that the efficacy of circRNA abnormal expression profile based on issue was equivalent to that of ACU based on plasma/serum; GAPDH based testing and non-GAPDH based testing could also obtain similar results (Table 5).
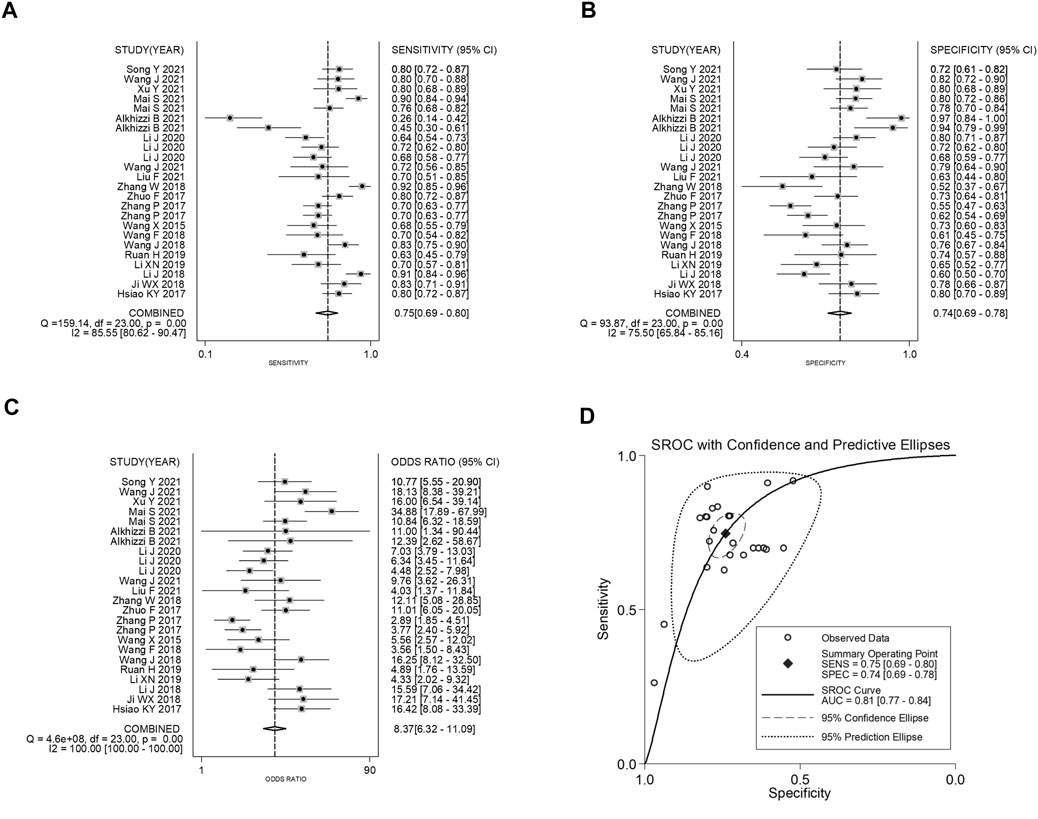
FIGURE 2. Forest plots of pooled (A) sensitivity, (B) specificity, (C) DOR, and (D) AUC for the diagnostic effect of circRNAs in CRC.
Prognostic Analysis
The subgroup classification was conducted according to the gene functions of circRNAs. The prognosis analysis showed that high expression levels ofup-regulated circRNAs were associated with the poor OS in GC patients (HR = 2.38, 95% CI: 1.66–3.41, p = 0.000), whereas the total survival time in GC patients with high expressed down-regulated circRNAs was significantly prolonged (an outlier eliminated adjusted HR = 0.33, 95% CI: 0.15–0.72, p = 0.006) (Figure 3).
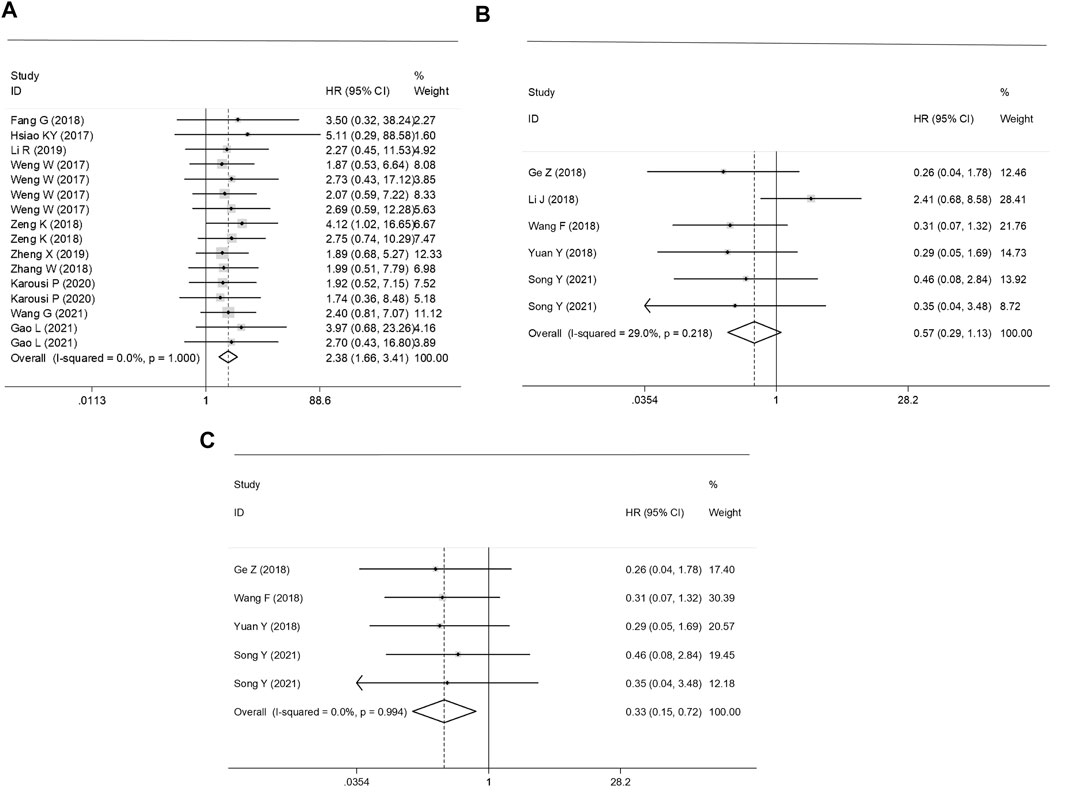
FIGURE 3. Forest plots of the prognostic effect of (A) up-regulated, (B) un-adjusted down-regulated circRNAs and (C) adjusted down-regulated circRNAs on survival in CRC patients.
Sensitivity Analysis and the Meta-Regression Test
Sources of heterogeneity were traced by a sensitivity analysis, and no outliers were identified in the diagnostic meta-analyses and the prognostic effect for the plasma/serum-based circRNA testing (Figures 4A–C). However, it was found that data of one independent study of the prognostic effect was identified as an outlier (18) (Figure 4D). After the removal of the outlier, the p value of Cochran’ Q test varied from 0.000 to 0.961, and I2 decreased to 0.0% from 29.0%. All this suggested that the inclusion of outliers for statistical consolidation could be considered as one of the important reasons for the heterogeneity of the analysis results. Thus, the adjusted HR of the down-regulated circRNAs was calculated after an elimination of the outlier (Figure 3C).
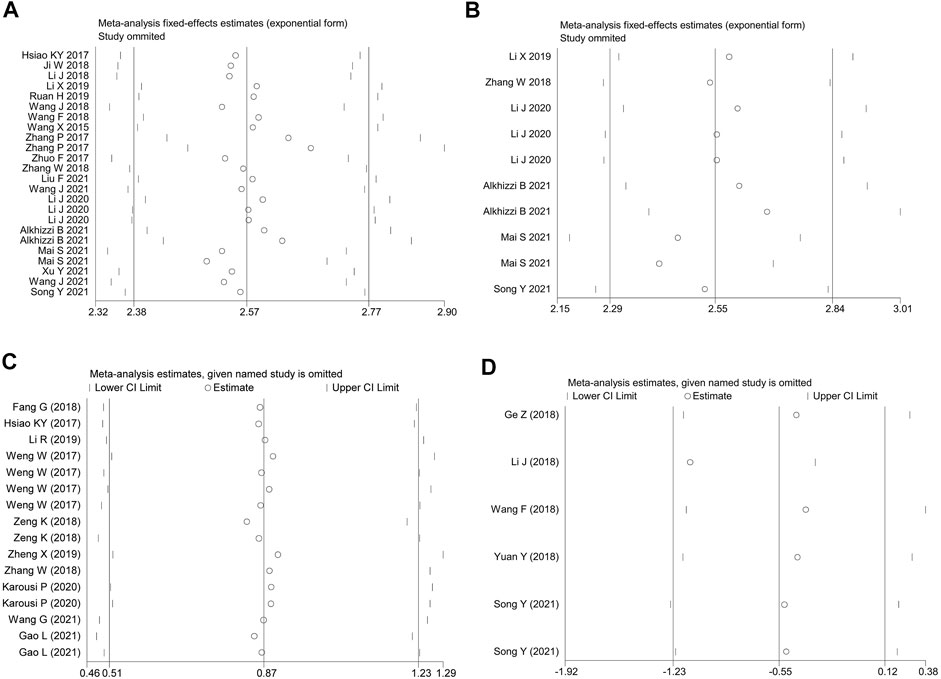
FIGURE 4. The influence analysis of the homogeneity among the included studies. (A) the overall diagnostic efficacy of the studies. (B) the diagnostic efficacy of plasma/serum-based circRNA testing. (C) up-regulated and (D) down-regulated circRNAs in predicting the OS in CRC patients.
On the other hand, a meta-regression test was performed to analyze factors comprising the quality of the study, the type of study, the type of specimen, the number of cases and the number of control groups. The results showed that these factors were not the source of heterogeneity among the studies with nonsignificant differences (data not shown).
Publication Bias
Assessment of the publication bias is shown in Figure 5. We observed publication bias in the combined prognostic effect forup-regulated circRNAs (Egger’s test, p = 0.021) (Figure 5C). The missing studies were adjusted through the trim-and-fill method (Figure 5D). Nevertheless, the pooled analysis incorporating the hypothetical studies (moment-based estimate of between studies variance = 0.000, p = 1.000) altered slightly from the unadjusted ones (moment-based estimate of between studies variance = 0.000, p = 1.000), suggesting that the bias did not have a significant impact on the combined effect ofup-regulated circRNAs in CRC.
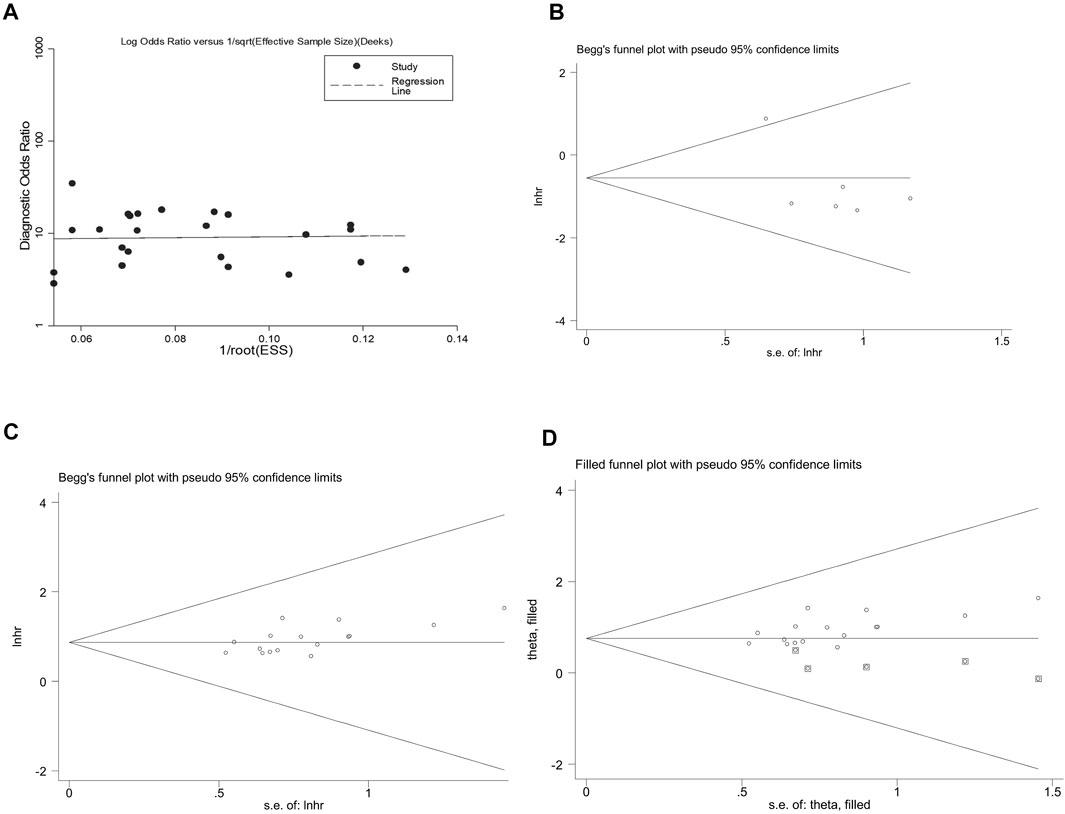
FIGURE 5. Appraising the publication bias among the included studies. (A) the overall diagnostic effect assessed by the Deek’s funnel plot. Begg’s test for the effects of (B) down-regulated and (C) up-regulated circRNAs in predicting the OS in CRC patients. (D) funnel plot of the trim-and-fill method shows no significant impacts on the combined effect of up-regulated circRNAs in CRC.
Discussion
CRC is a common malignant tumor of the digestive tract, with nearly 0.6 million newly diagnosed cases across the world every year (1,2). Early detection, diagnosis and treatment of the disease can maximize the survival of the patients (3,5,7). CircRNAs as a class of coding/non-coding competitive endogenous RNA, are characterized by a closed covalent continuous ring structure without the 5′-3′ structure and a poly (A) tail (8,9). A large number of circRNAs existing in mammals affect their functional expressions by binding to miRNAs and other target molecules at the transcriptional or post-transcriptional level (10,11). Although mechanisms behind their biofunctions have not been completed expounded, increasing evidences have shown that circRNAs are associated with multiple diseases, especially malignant tumors (12,13). In recent years, it has been found that expression profiles of circRNAs in CRC cancerous tissues are significantly different from those in the corresponding paracancerous tissues, suggesting that the abnormally expressed circRNAs can be critical in the onset and development of this malady. And they are expected to become new biomarkers in the early diagnosis and treatment of CRC (14–31). Herein, we conducted a pair-wire meta-analysis, and assessed the diagnostic and prognostic significance of abnormally expressed circRNAs in CRC.
In our study, expression levels of circRNAs present clinical values in its diagnosis, with the sensitivity and specificity of 0.75 and 0.74 respectively and the corresponding AUC of 0.81. DOR, the rate of true positive to false positive, is another important indice to evaluate the effectiveness of circRNA-based diagnostic tests (46). A DOR of less than 1 indicates the very low efficiency in the diagnosis of CRC (46). However, a prominent outcome of 8.37 was presented in our study, suggesting the high efficiency of circRNA-based diagnosis. In addition, the pooled PLR of 2.86 means the probability of positive circRNA expressions in CRC patients is approximately 5 times higher than that in controls. The pooled NLR of 0.34 indicates that only 34% of the negative results of circRNA tests are false negative. These findings fully prove that circRNA detection can be applied in the early diagnosis of CRC. As proven by two previous studies, the sensitivity, specificity and AUC of combined circRNA expression profiles in the diagnosis of all malignant tumors are 0.72, 0.74, and 0.79 (47), and 0.79, 0.73, and 0.83 in the diagnosis of gastrointestinal cancers (48). These findings are consistent with our results.
The expression characteristics and sample types of circRNAs have been analyzed by subgroup analyses. We find that up-regulated circRNAs are more effective in diagnosing CRC than down-regulated circRNAs, acting as oncogenes. The analysis based on the sample type indicates that the high kurtosis in the newly diagnosed CRC patients is more beneficial to the detection. The AUC in tissue-derived circRNAs is higher than that in plasma-derived ones, suggesting that the tissue-based circRNA tests may be more accurate than the plasma-related tests; however, this also may attribute to a higher expression peak of circRNAs in cancerous tissues. In spite of this, the number of samples in the subgroup analysis is lower than that of the whole, and the conclusion needs to be confirmed by large sample size trials with later-stage patients.
As circRNAs have been proven to be associated with the prognosis of malignant tumors, systematical evaluations for the prognostic efficacy of circRNA expressions in lung cancer and hepatocellular carcinoma have been conducted (49,50). The results show that the higher the levels of up-regulated circRNAs are, the worse the prognosis of the cancer patients will be. Evidences show that expressions of down-regulated circRNAs in hepatocellular carcinoma are associated with a dismal survival time. These patients with high expressed circRNAs are more likely to extend the OS time. It can be seen that biological functions of different expression states of circRNAs in malignant tumors are distinct. Therefore, we evaluated circRNA as a potential prognostic biomarker of CRC. According to the principle of different biofunctions of circRNAs corresponding to their types, we have divided the circRNA expression profile into two groups: oncogenes and tumor-suppressor ones. The survival analysis showed that the total survival time of the patients with high-expressed up-regulated circRNAs was significantly shorter than that of patients with low-expression levels. And the CRC patients with low expressions of up-regulated circRNAs and high expressions of tumor-suppressor circRNAs significantly presented favorable prognosis, suggesting that circRNA profiling can be used as indicators for evaluating and monitoring the prognosis of CRC.
The main sources of heterogeneity in a meta-analysis consist of the threshold effect and the non-threshold effect. Results of the Spearman correlation coefficient analysis indicate that the heterogeneity in the overall merger statistics and subgroup analyses mainly result from the threshold effect, which may attribute to different boundary values or cut-off values. In this study, the relative quantitative cut-off values and internal reference genes for circRNAs are different, which can be one of the main causes of heterogeneity. Besides, our study has discussed the possible factors leading to heterogeneity by the sensitivity analysis and the meta-regression analysis. We identified an outlier study in the sensitivity analysis and its impact on the prognostic effect was verified. However, the meta-regression test has ruled out factors including the quality of the study, the type of study, the type of specimen, the number of cases and the number of control groups as the possible sources of heterogeneity among the studies.
As the biofunctions of circRNAs in CRC have been clarified above, some limitations still exist in our study. Firstly, the types of circRNA molecules and samples enrolled are not uniform, featuring a large heterogeneity among the studies. Secondly, the included subjects are predominantly Chinese population, which means certain population bias exists in the merged data. Thirdly, there are merely a handful of studies using plasma- or serum-derived circRNAs, and the relevant findings need to be verified by further researches.
Conclusion
To sum up, circRNAs can be used as promising auxiliary biomarkers for the diagnosis and prognosis prediction of CRC. Our findings still need to be confirmed based on more high-quality trials.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
CL was in charge of designing, writing and supervising whole process; Q-BX was in charge of collecting date; LD was in charge of analyzing date; L-JH was in charge of decorating words; YJ is the corresponding author, in charge of all process as well.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel, RL, Miller, KD, Fuchs, HE, and Jemal, A. Cancer Statistics, 2021. CA A Cancer J Clin (2021) 71(1):7–33. PMID: 33433946. doi:10.3322/caac.21654
2. Chen, W, Zheng, R, Baade, PD, Zhang, S, Zeng, H, Bray, F, et al. Cancer Statistics in China, 2015. CA: a Cancer J clinicians (2016) 66(2):115–32. PMID: 26808342. doi:10.3322/caac.21338
3. Simon, K. Colorectal Cancer Development and Advances in Screening. Clin Interv Aging (2016) 11:967–76. PMID: 27486317. doi:10.2147/CIA.S109285
4. Díaz-Tasende, J. Colorectal Cancer Screening and Survival. Rev Esp Enferm Dig (2018) 110(11):681–3. PMID: 30284905. doi:10.17235/reed.2018.5870/2018
5. Maida, M, Macaluso, FS, Ianiro, G, Mangiola, F, Sinagra, E, Hold, G, et al. Screening of Colorectal Cancer: Present and Future. Expert Rev Anticancer Ther (2017) 17(12):1131–46. PMID: 29022408. doi:10.1080/14737140.2017.1392243
6. Dumoulin, FL, and Hildenbrand, R. Endoscopic Resection Techniques for Colorectal Neoplasia: Current Developments. Wjg (2019) 25(3):300–7. PMID: 30686899. doi:10.3748/wjg.v25.i3.300
7. Nicholson, BD, Shinkins, B, Pathiraja, I, Roberts, NW, James, TJ, Mallett, S, et al. Blood CEA Levels for Detecting Recurrent Colorectal Cancer. Cochrane Database Syst Rev (2015) 2015(12):CD011134. PMID: 26661580. doi:10.1002/14651858.CD011134.pub2
8. Fischer, JW, and Leung, AKL. CircRNAs: a Regulator of Cellular Stress. Crit Rev Biochem Mol Biol (2017) 52(2):220–33. PMID: 28095716. doi:10.1080/10409238.2016.1276882
9. Patop, IL, Wüst, S, and Kadener, S. Past, Present, and Future of Circ RNA S. EMBO J (2019) 38(16):e100836. PMID: 31343080. doi:10.15252/embj.2018100836
10. Wilusz, JE. A 360° View of Circular RNAs: From Biogenesis to Functions. Wiley Interdiscip Rev RNA (2018) 9(4):e1478. PMID: 29655315. doi:10.1002/wrna.1478
11. Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet (2016) 32(5):309–16. [PMID: 27050930]. doi:10.1016/j.tig.2016.03.002
12. Kristensen, LS, Hansen, TB, Venø, MT, and Kjems, J. Circular RNAs in Cancer: Opportunities and Challenges in the Field. Oncogene (2018) 37(5):555–65. PMID: 28991235. doi:10.1038/onc.2017.361
13. Meng, S, Zhou, H, Feng, Z, Xu, Z, Tang, Y, Li, P, et al. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol Cancer (2017) 16(1):94. PMID: 28535767. doi:10.1186/s12943-017-0663-2
14. Fang, G, Ye, B-L, Hu, B-R, Ruan, X-J, and Shi, Y-X. CircRNA_100290 Promotes Colorectal Cancer Progression through miR-516b-Induced Downregulation of FZD4 Expression and Wnt/β-Catenin Signaling. Biochem Biophysical Res Commun (2018) 504(1):184–9. PMID: 30173892. doi:10.1016/j.bbrc.2018.08.152
15. Ge, Z, Li, LF, Wang, CY, Wang, Y, and Ma, WL. CircMTO1 Inhibits Cell Proliferation and Invasion by Regulating Wnt/β-Catenin Signaling Pathway in Colorectal Cancer. Eur Rev Med Pharmacol Sci (2018) 22(23):8203–9. PMID: 30556859. doi:10.26355/eurrev_201812_16513
16. Hsiao, K-Y, Lin, Y-C, Gupta, SK, Chang, N, Yen, L, Sun, HS, et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res (2017) 77(9):2339–50. PMID: 28249903. doi:10.1158/0008-5472.CAN-16-1883
17. Ji, W, Qiu, C, Wang, M, Mao, N, Wu, S, and Dai, Y. Hsa_circ_0001649: A Circular RNA and Potential Novel Biomarker for Colorectal Cancer. Biochem Biophysical Res Commun (2018) 497(1):122–6. PMID: 29421663. doi:10.1016/j.bbrc.2018.02.036
18. Li, J, Ni, S, Zhou, C, and Ye, M. The Expression Profile and Clinical Application Potential of Hsa_circ_0000711 in Colorectal Cancer. Cmar (2018) Vol. 10:2777–84. PMID: 30147374. doi:10.2147/cmar.s172388
19. Li, R, Wu, B, Xia, J, Ye, L, and Yang, X. Circular RNA hsa_circRNA_102958 Promotes Tumorigenesis of Colorectal Cancer via miR-585/CDC25B axis. Cmar (2019) Vol. 11:6887–93. PMID: 31413634. doi:10.2147/cmar.s212180
20. Li, X-N, Wang, Z-J, Ye, C-X, Zhao, B-C, Huang, X-X, and Yang, L. Circular RNA circVAPA Is Up-Regulated and Exerts Oncogenic Properties by Sponging miR-101 in Colorectal Cancer. Biomed Pharmacother (2019) 112:108611. PMID: 30797148. doi:10.1016/j.biopha.2019.108611
21. Ruan, H, Deng, X, Dong, L, Yang, D, Xu, Y, Peng, H, et al. Circular RNA Circ_0002138 Is Down-Regulated and Suppresses Cell Proliferation in Colorectal Cancer. Biomed Pharmacother (2019) 111:1022–8. PMID: 30841415. doi:10.1016/j.biopha.2018.12.150
22. Wang, F, Wang, J, Cao, X, Xu, L, and Chen, L. Hsa_circ_0014717 Is Downregulated in Colorectal Cancer and Inhibits Tumor Growth by Promoting P16 Expression. Biomed Pharmacother (2018) 98:775–82. PMID: 29571246. doi:10.1016/j.biopha.2018.01.015
23. Wang, J, Li, X, Lu, L, He, L, Hu, H, and Xu, Z. Circular RNA Hsa_circ_0000567 Can Be Used as a Promising Diagnostic Biomarker for Human Colorectal Cancer. J Clin Lab Anal (2018) 32(5):e22379. PMID: 29333615. doi:10.1002/jcla.22379
24. Wang, X, Zhang, Y, Huang, L, Zhang, J, Pan, F, Li, B, et al. Decreased Expression of Hsa_circ_001988 in Colorectal Cancer and its Clinical Significances. Int J Clin Exp Pathol (2015) 8(12):16020–5. PMID: 26884878.
25. Weng, W, Wei, Q, Toden, S, Yoshida, K, Nagasaka, T, Fujiwara, T, et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res (2017) 23(14):3918–28. PMID: 28174233. doi:10.1158/1078-0432.CCR-16-2541
26. Yuan, Y, Liu, W, Zhang, Y, Zhang, Y, and Sun, S. CircRNA Circ_0026344 as a Prognostic Biomarker Suppresses Colorectal Cancer Progression via microRNA-21 and microRNA-31. Biochem Biophysical Res Commun (2018) 503(2):870–5. PMID: 29928882. doi:10.1016/j.bbrc.2018.06.089
27. Zeng, K, Chen, X, Xu, M, Liu, X, Hu, X, Xu, T, et al. CircHIPK3 Promotes Colorectal Cancer Growth and Metastasis by Sponging miR-7. Cell Death Dis (2018) 9(4):417. PMID: 29549306. doi:10.1038/s41419-018-0454-8
28. Zhang, P, Zuo, Z, Shang, W, Wu, A, Bi, R, Wu, J, et al. Identification of Differentially Expressed Circular RNAs in Human Colorectal Cancer. Tumour Biol (2017) 39(3):101042831769454–1010428317694546. PMID: 28349836. doi:10.1177/1010428317694546
29. Zhang, W, Yang, S, Liu, Y, Wang, Y, Lin, T, Li, Y, et al. Hsa_circ_0007534 as a Blood-Based Marker for the Diagnosis of Colorectal Cancer and its Prognostic Value. Int J Clin Exp Pathol (2018) 11(3):1399–406. PMID: 31938236.
30. Zheng, X, Chen, L, Zhou, Y, Wang, Q, Zheng, Z, Xu, B, et al. A Novel Protein Encoded by a Circular RNA circPPP1R12A Promotes Tumor Pathogenesis and Metastasis of colon Cancer via Hippo-YAP Signaling. Mol Cancer (2019) 18(1):47. PMID: 30925892. doi:10.1186/s12943-019-1010-6
31. Zhuo, F, Lin, H, Chen, Z, Huang, Z, and Hu, J. The Expression Profile and Clinical Significance of circRNA0003906 in Colorectal Cancer. Ott (2017) Vol. 10:5187–93. PMID: 29123417. doi:10.2147/ott.s147378
32. Liu, F, Xiao, X-L, Liu, Y-J, Xu, R-H, Zhou, W-J, Xu, H-C, et al. CircRNA_0084927 Promotes Colorectal Cancer Progression by Regulating miRNA-20b-3p/glutathione S-Transferase Mu 5 axis. Wjg (2021) 27(36):6064–78. PMID: 34629820 PMCID: PMC8476332. doi:10.3748/wjg.v27.i36.6064
33. Wang, J, Wang, T, Hu, S, Li, J, Ni, C, and Ye, M. Hsa_circ_0043278 Inhibits Tumorigenesis and Is Downregulated in Colorectal Cancer. Cmar (2021) Vol. 13:965–75. PMID: 33568942 PMCID: PMC7868253. doi:10.2147/cmar.s289775
34. Li, J, Song, Y, Wang, J, and Huang, J. Plasma Circular RNA Panel Acts as a Novel Diagnostic Biomarker for Colorectal Cancer Detection. Am J Transl Res (2020) 12(11):7395–403. PMID: 33312376 PMCID: PMC7724351.
35. Alkhizzi, B, Khan, M, Al-Ghafari, A, and Choudhry, H. Upregulation of Circular and Linear METTL3 and USP3 in Colorectal Cancer. Oncol Lett (2021) 22(3):675. PMID: 34345300 PMCID: PMC8323003. doi:10.3892/ol.2021.12936
36. Mai, S, Zhang, Z, and Mi, W. Upregulation of circ_PVT1 and Circ_001569 Indicate Unfavorable Prognosis in Colorectal Cancer. Ann Clin Lab Sci (2021) 51(1):55–60. PMID: 33653781.
37. Xu, Y, Wang, C, Lu, X, Qi, R, Wang, X, and Zhao, J. CircRNA_002178 as a ceRNA Promotes the Development of Colorectal Cancer by Regulating miR-542-3p/CREB1. Am J Transl Res (2021) 13(9):10038–55. PMID: 34650680 PMCID: PMC8507087.
38. Wang, J, Zhang, Y, Song, H, Yin, H, Jiang, T, Xu, Y, et al. The Circular RNA circSPARC Enhances the Migration and Proliferation of Colorectal Cancer by Regulating the JAK/STAT Pathway. Mol Cancer (2021) 20(1):81. PMID: 34074294 PMCID: PMC8167978. doi:10.1186/s12943-021-01375-x
39. Song, Y, Cao, P, and Li, J. Plasma Circular RNA Hsa_circ_0001821 Acts as a Novel Diagnostic Biomarker for Malignant Tumors. J Clin Lab Anal (2021) 35(11):e24009. PMID: 34523755. doi:10.1002/jcla.24009
40. Karousi, P, Artemaki, PI, Sotiropoulou, CD, Christodoulou, S, Scorilas, A, and Kontos, CK. Identification of Two Novel Circular RNAs Deriving from BCL2L12 and Investigation of Their Potential Value as a Molecular Signature in Colorectal Cancer. Ijms (2020) 21(22):8867. PMID: 33238574 PMCID: PMC7709015. doi:10.3390/ijms21228867
41. Wang, G, and Yang, H. CircRNA DUSP16 Knockdown Suppresses Colorectal Cancer Progression by Regulating the miR-432-5p/E2F6 Axis. Cmar (2021) Vol. 13:6599–609. PMID: 34456589 PMCID: PMC8387644. doi:10.2147/cmar.s323437
42. Gao, L, Tang, X, He, Q, Sun, G, Wang, C, and Qu, H. Exosome-transmitted circCOG2 Promotes Colorectal Cancer Progression via miR-1305/tgf-Β2/smad3 Pathway. Cell Death Discov. (2021) 7(1):281. PMID: 34635639 PMCID: PMC8505430. doi:10.1038/s41420-021-00680-0
43. Whiting, PF, Rutjes, AWS, Westwood, ME, Mallett, S, Deeks, JJ, Reitsma, JB, et al. QUADAS-2 Group. QUADAS-2: a Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med (2011) 155(8):529–36. PMID: 22007046. doi:10.7326/0003-4819-155-8-201110180-00009
44. Stang, A. Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur J Epidemiol (2010) 25(9):603–5. PMID: 20652370. doi:10.1007/s10654-010-9491-z
45. Lin, N, Lin, Y, Fu, X, Wu, C, Xu, J, Cui, Z, et al. MicroRNAs as a Novel Class of Diagnostic Biomarkers in Detection of Oral Carcinoma: a Meta-Analysis Study. Clin Lab (2016) 62(3):451–61. PMID: 27156336. doi:10.7754/clin.lab.2015.150802
46. Glas, AS, Lijmer, JG, Prins, MH, Bonsel, GJ, and Bossuyt, PMM. The Diagnostic Odds Ratio: a Single Indicator of Test Performance. J Clin Epidemiol (2003) 56(11):1129–35. PMID: 14615004. doi:10.1016/s0895-4356(03)00177-x
47. Wang, M, Yang, Y, Xu, J, Bai, W, Ren, X, and Wu, H. CircRNAs as Biomarkers of Cancer: a Meta-Analysis. BMC Cancer (2018) 18(1):303. PMID: 29554887. doi:10.1186/s12885-018-4213-0
48. Lin, Y, Luo, X, Cui, Z, and Chen, Y. Diagnostic Potential for Circular RNAs in Gastric Carcinoma: a Meta-Analysis. Clin Lab (2019) 65(3). PMID: 30868864. doi:10.7754/Clin.Lab.2018.180810
49. Hao, Q, Han, Y, Xia, W, Wang, Q, and Qian, H. Systematic Review and Meta-Analysis of the Utility of Circular RNAs as Biomarkers of Hepatocellular Carcinoma. Can J Gastroenterol Hepatol (2019) 2019:1–13. PMID: 31187026. doi:10.1155/2019/1684039
Keywords: colorectal cancer, circular RNA, diagnosis, prognosis, pooled analysis
Citation: Long C, Xu Q-b, Ding L, Huang L-j and Ji Y (2022) Circular RNAs as Diagnostic and Prognostic Indicators of Colorectal Cancer: A Pooled Analysis of Individual Studies. Pathol. Oncol. Res. 28:1610037. doi: 10.3389/pore.2022.1610037
Received: 23 August 2021; Accepted: 08 February 2022;
Published: 17 March 2022.
Edited by:
Andrea Ladányi, National Institute of Oncology (NIO), HungaryCopyright © 2022 Long, Xu, Ding, Huang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Ji, ampybXlvbmdqaUAxNjMuY29t
 Cong Long
Cong Long Qiu-bo Xu1
Qiu-bo Xu1