Abstract
Cells of the monocyte macrophage lineage form multinucleated giant cells (GCs) by fusion, which may express some cell cycle markers. By using a comprehensive marker set, here we looked for potential replication activities in GCs, and investigated whether these have diagnostic or clinical relevance in giant cell tumor of bone (GCTB). GC rich regions of 10 primary and 10 first recurrence GCTB cases were tested using immunohistochemistry in tissue microarrays. The nuclear positivity rate of the general proliferation marker, replication licensing, G1/S-phase, S/G2/M-phase, mitosis promoter, and cyclin dependent kinase (CDK) inhibitor reactions was analyzed in GCs. Concerning Ki67, moderate SP6 reaction was seen in many GC nuclei, while B56 and Mib1 positivity was rare, but the latter could be linked to more aggressive (p = 0.012) phenotype. Regular MCM6 reaction, as opposed to uncommon MCM2, suggested an initial DNA unwinding. Early replication course in GCs was also supported by widely detecting CDK4 and cyclin E, for the first time, and confirming cyclin D1 upregulation. However, post-G1-phase markers CDK2, cyclin A, geminin, topoisomerase-2a, aurora kinase A, and phospho-histone H3 were rare or missing. These were likely silenced by upregulated CDK inhibitors p15INK4b, p16INK4a, p27KIP1, p53 through its effector p21WAF1 and possibly cyclin G1, consistent with the prevention of DNA replication. In conclusion, the upregulation of known and several novel cell cycle progression markers detected here clearly verify early replication activities in GCs, which are controlled by cell cycle arresting CDK inhibitors at G1 phase, and support the functional maturation of GCs in GCTB.
Introduction
There are two major ways of forming multinucleate giant cells i.e. acytokinetic cell division and cell fusion [1]. Proliferating neoplastic cells, e.g. Reed-Sternberg cells in Hodgkin’s lymphoma or multinucleated tumor cells in soft tissue, e.g. myxofibro- and osteosarcomas are resulted from incomplete cell division as a result of their cytoskeleton vulnerability e.g. of the contractile ring [2]. On the contrary, inflammatory multinucleated giant cells (GC), such as osteoclast-type giant cells, Langhans-type granuloma giant cells and foreign body giant cells are formed by fusion of cells of the monocyte-macrophage lineage [3].
By testing the expression of cell cycle phase progression associated markers in the mononuclear cells of GCTB, we recently showed that cases with elevated post-G1-phase cell fraction, indicating accelerated cell cycle progression, may predict shorter progression free survival (PFS) [4]. We also recognized that GC nuclei may show diverse proportion of immunoreactions not only for the earlier detected cyclin D and p21WAF1 [5, 6], and cell cycle control proteins, but also for some cell cycle licensing and promoter markers, which had not been noticed before, despite GCs are considered to be of reactive, osteoclastic phenotype [7]. Therefore, here we studied the expression of a comprehensive set of cell cycle regulatory proteins to see if GCs in GCTB are still show replicative activity and if it has a clinico-pathological relevance.
GCTB is an osteolytic, locally destructive bone lesion, which, besides GCs, is made up mainly of mononuclear monocytic cells which act as precursors for GCs, and of neoplastic stromal cells (Figure 1). The proliferating, neoplastic stromal cells, generally carrying H3F3A G34W mutation [8], are the major drivers of osteoclastogenesis and pathological bone resorption [9, 10]. They produce canonical (RANKL/M-CSF) and non-canonical (e.g., LIGHT, TNFα, IL-6 or vascular endothelial growth factor–VEGF) growth factors and hypoxia inducible factors 1α and 2α [14], which can either directly or through autocrine activation promote osteoclastogenesis and osteolysis [11, 12]. We have shown earlier that besides the replication activity of neoplastic stromal cells, their elevated epidermal growth factor receptor (EGFR) signaling and deregulated gap junction connexin43 expression and channel functions, can contribute to GCTB progression, mediated by GCs [4, 13, 14].
FIGURE 1
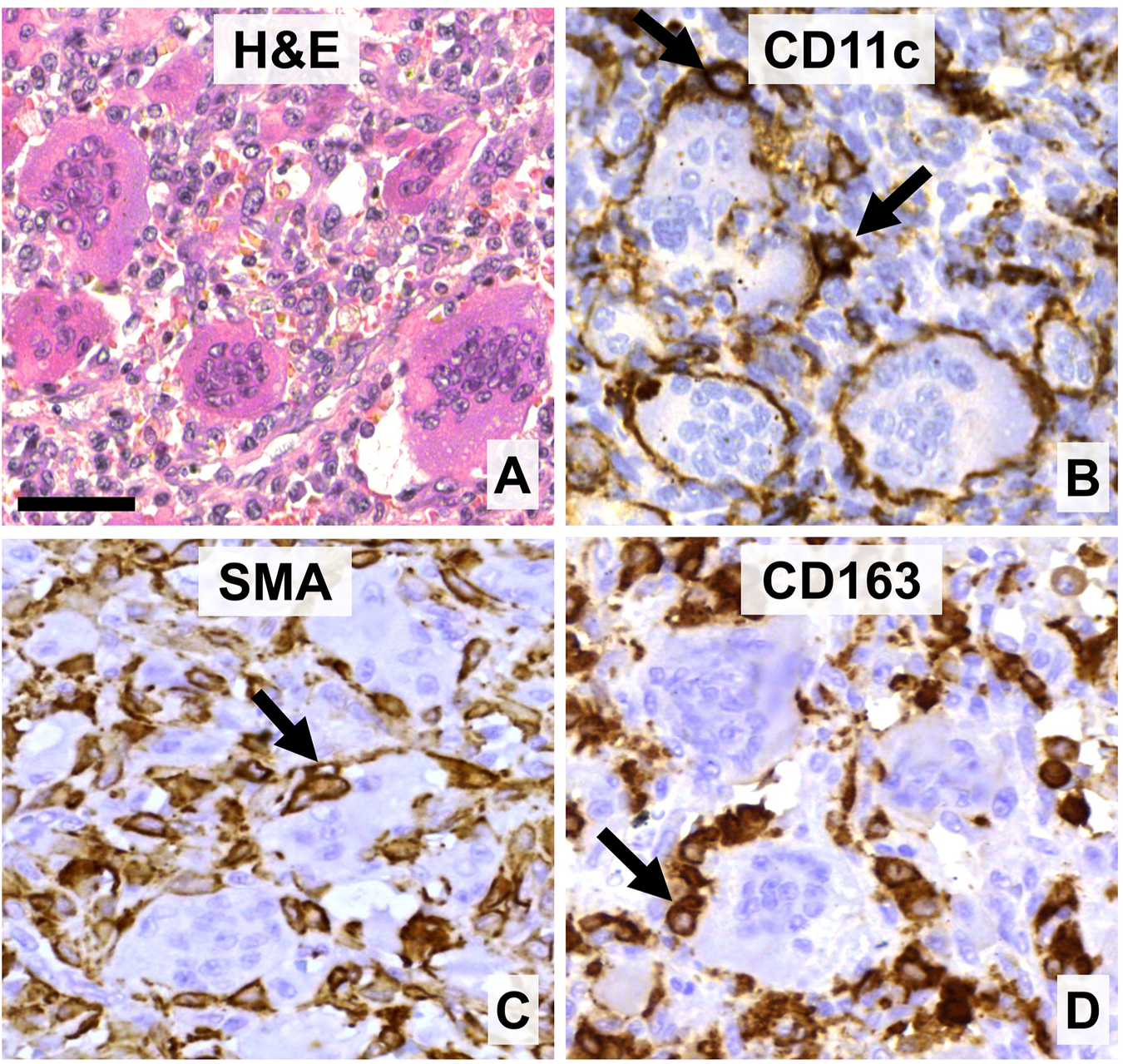
The main cell components of giant cell tumor of bone. (A) Multinucleated giant cells (GCs) of different sizes are intermingled with mononuclear and red blood cells (hematoxylin-eosin staining, (H&E). (B) GCs and their monocytic precursors are positive for CD11c. (C) Neoplastic stromal cells can be positive for smooth muscle actin (SMA). (D) Histiocytes and monocytic GC precursors but not GCs express CD163 scavenger receptor. Arrows show immunopositive cells intimate association with GCs. DAB (brown) immunoperoxidase reactions (B–D). Scale bar: 50 μm for all images.
In the present study, we tested the expression of cell cycle regulatory proteins in GC nuclei, including 3 clones for the general proliferation marker Ki67; the DNA replication licensing factors MCM2 and MCM6; the G1-S phase marker cyclin D1 and its complexing partner CDK4/6; the early (CDK2 and cyclin A) and late (topoisomerase 2–TOP2) post-G1 phase markers; and the G2-M-phase markers (aurora kinase A–AURKA, and phospho-histone-H3–pHH3) [15]. Furthermore, the DNA replication inhibitor geminin, the cyclin dependent kinase inhibitors p15INK4b, p16INK4a, p21WAF1 and p27KIP1; as well as the oncosuppressor retinoblastoma and p53 [16], and the unpaired cyclin G1 [17, 18] proteins were also examined. We aimed to profile cell cycle activity in GCs and see if it is different between primary and recurrent GCTB.
Materials and Methods
Study Cohort
This study was performed on immunostained 2 mm diameter, 70 sample tissue microarray (TMA) sections of formalin-fixed and paraffin-embedded samples of GCTB cases, which were diagnosed between 1994–2005 at the Institute of Rizzoli, Bologna (IOR), Italy (ethical approval: IOR 13351/5-28-2008 and Semmelweis University: 87/2007) [9,10]. A stratified random sample of 10 primary tumors (P) and 10 first recurrences (1-Rec) were selected from our previously published single-center retrospective study [9]. The clinicopathological characteristics of the selected cohort are presented in Table 1. Briefly, of the 20 patients, 12 progression events were registered (60%) during follow-up. During the study period 12 progression events were registered. Eight-eight patients (40%) were continuously disease free or had local recurrences respectively, 2 (10%) were alive with disease at last follow-up while malignant transformation and stroke both with consecutive fatal outcomes occurred in 1-1 patients (5%).
TABLE 1
| Number of patients | 20 |
|---|---|
| Number of surgical cases | 20 |
| Progression groups | Nr. Enneking’s/Campanacci’s grade |
| Primary | 10 L 3 A 5 Ag 2 |
| 1st recurrent cases | 10 L 2 A 4 Ag 4 |
| Median age (at case diagnosis) | 30.8 years (range: 13.7–75.6 years) |
| Sex (female, %) | 13 (65) |
| Sex ratio | 0.54:1 (m/f) or 1:1.85 (f/m) |
| Survival | |
| Median recurrence survival | 58.1 months (range:5.5–159.5, IQR: 18.9–79.2) |
| Number of progression events | 12 |
| Localization | Total (%) |
| Upper limb | 5 (25) |
| Lower limb | 13 (65) |
| Central (Sacrum + Spine or Pelvis) | 2 (10) |
| Treatment types | Total (%) |
| Curettage | 12 (60) |
| Resection or Amputation or Excision | 6 (30) |
| Radiotherapy | 2 (10) |
Clinicopathological features of the study cohort.
GCTB, giant cell tumor of bone; L, latent; A, active; Ag, aggressive; LQ/UQ, lower/upper quartile
Immunohistochemistry
Following routine dewaxing, the antigen retrieval of TMA sections was done in an electric pressure cooker (Avair, Bitalon, Pecs, Hungary) using 0.01 M Tris–0.1 M EDTA (TE) at ∼105°C for 30 min. Mouse or rabbit monoclonal primary antibody clones, or rabbit polyclonal immunoglobulins were incubated overnight (16 h) at room temperature. These included anti-Ki67 Mib1 (1:100; Dako, Glostrup, Denmark), B56 (1:100; Histopathology, Pecs, Hungary), SP6 (1:600); anti-MCM2 (CRCT2.1, 1:200) and -MCM6 (1:600, PA5-79649); anti-cyclin D1 (SP4, 1:200), -CDK2 (1:300, 2B6), -CDK4 (1:300, DCS31 + 35), -cyclin E (13A3, 1:20), -cyclin G (11C8, 1:100), -cyclin A (6E6, 1:500), -topoisomerase 2 (Ki-S1, 1:200), -aurora kinase A (1G4, 1:80; Cell Signaling, Danvers, MA, United States), -pHH3Ser10 (K872.3, 1:100); -p53 (DO7, 1:100), -retinoblastoma (1F8, 1:100), -p15INK4b (15P06, 1:200), -p16INK4a (JC8, 1:200), -p21WAF1 (SX118, 1:100), and -geminin (EM6, 1:150; Leica-NovoCastra, Newcastle Upon-Tyne, United Kingdom), -p53 (DO7, 1:200, Leica-NovoCastra; and BP53–12, 1:100), and -retinoblastoma (51B7, 1:100) immunoglobulins (IgGs). Except where otherwise indicated, all antibodies were from Thermo-Fisher LabVision (Fremount, CA, United States). Then, the NovoLink polymer peroxidase kit (Leica-NovoCastra) was used as a detection system for 60 min. Immunoreactions were revealed by using DAB Quatro kit (Thermo-Fisher) for 3–5 min under microscopic control and the sections were coverslip mounted after hematoxylin nuclear conterstaining. For double immunofluorescence (C) mouse Ki67 (Mib1, green) and rabbit cyclin D1 (SP4, red) antibodies, were detected simultaneously using Alexa Fluor 488 goat anti-mouse IgG (1:200, green; code: A11001), Alexa Fluor 564 goat anti-rabbit IgG (1:200, red; code: A11035). The immunostained TMA sections were digitalized with the Pannoramic® Scan II System and analyzed using its CaseViewer software (3DHISTECH, Budapest, Hungary).
Evaluation of the Immunoreactions
After setting up and agreeing on the evaluation criteria with the project leader (TK), systematic assessment was done by an independent assessor (MEM) blinded to all clinical- and other cell cycle marker expression data in three different osteoclast/giant cell rich high-power fields (HPF; 80x) in each case (overall ∼1000 regions of interest) [4]. All nuclear positivity, which was obvious compared to adjacent negative cells showing only hematoxylin staining, was counted. Additional cytoplasmic staining was considered only for p16INK4a on a four-grade Likert scale (0-negative, 1-weak, 2-moderate, 3-strong). The number of GCs (NGC), GC nuclei (NGC_nuclei) and respective cell cycle marker positive GC nuclei (NGC_nuclei+) were recorded and averaged for each case. To robustly estimate NGC and NGC_nuclei in a surgical specimen, their values were averaged over all tested cell cycle markers for each case, respectively. As NGC_nuclei+ values are dependent on multiple factors e.g. the position of the TMA core within the specimen, NGC and properties of the CC marker, their absolute values showed substantial variances and were not necessarily comparable across different immunostainings. Therefore, we normalized these values by calculating their ratio for each staining (NGC_nuclei+/NGC_nuclei) to allow for more stable and direct comparisons across cell cycle markers.
Statistical Analyses
All analyses were performed with the R statistics program (v.3.6.3, R Core Team 2020, Vienna Austria; RStudio IDE v. 1.2.5033, Boston, MA, United States). Non-normally distributed variables were displayed as median, range and interquartile range (IQR). Categorical variables were reported as proportions. The Jonckheere–Terpstra test was used to investigate the overall difference between Enneking’s/Campanacci’s grading (i.e. latent, active and aggressive), GC count and GC nuclear positivity. We used the nonparametric Wilcoxon-Mann-Whitney U test for two samples comparing the mean rank of NGC, NGC_nuclei, and NGC_nuclei+ as well as their ratios between primary and recurrent samples [19]. Uni- and multivariate Cox proportional hazards models of time-to-first-event analyses were performed to explore possible associations between NGC_nuclei+ and progression free survival (PFS) [4]. Figures were generated with the ggplot2 library using colorblind-friendly palettes. p-values were adjusted for multiple testing to counteract type 1 error inflation using the conservative Bonferroni correction. Adjusted p-values (p*) <0.05 were considered significant.
Results
General Proliferation Markers in Giant Cells
Based on the potential importance of GC functions in GCTB progression, which may be linked to proliferation, first we counted the number and nuclear density of GCs. Though, neither the overall average GC number (NGC; W = 59, p = 0.53) nor the average number of GC nuclei (NGC_nuclei; W = 49, p = 0.97) showed statistical difference between P and 1-Rec GCTB cases, there was a trend of inverse relationship between the radiological grade (latent: L; active: A; aggressive: Ag) of GCTB and the overall average NGC (WL_vs_Ag = 30, pL_vs_Ag = 0.065; WA_vs_Ag = 37, pA_vs_Ag = 0.11) and NGC_nuclei (WL_vs_A = 29, pL_vs_A = 0.093). The distribution of the ratio of cell cycle marker positive GC nuclei in primary and 1-Rec GCTB cases is summarized in Table 2.
TABLE 2
| Type of material | Marker | Ratio of positive GC nuclei | Statistic (W) | p | p*Bonferroni (n = 14) | padj | |||
|---|---|---|---|---|---|---|---|---|---|
| Median | IQR | min | Max | ||||||
| P | CDK2 | 0.031 | 0.055 | 0 | 0.111 | 72 | 0.10 | 0.0036 | n.s. |
| 1-Rec | CDK2 | 0.004 | 0.012 | 0 | 0.078 | ||||
| P | CDK4 | 0.325 | 0.642 | 0 | 0.95 | 28 | 0.72 | 0.0036 | n.s. |
| 1-Rec | CDK4 | 0.412 | 0.193 | 0.043 | 0.826 | ||||
| P | Cyclin A | 0 | 0 | 0 | 0 | 40 | 0.17 | 0.0036 | n.s. |
| 1-Rec | Cyclin A | 0 | 0 | 0 | 0.006 | ||||
| P | Cyclin D1 | 0.941 | 0.13 | 0.694 | 0.994 | 66 | 0.25 | 0.0036 | n.s. |
| 1-Rec | Cyclin D1 | 0.874 | 0.152 | 0.462 | 0.981 | ||||
| P | Cyclin G1 | 1 | 0.018 | 0.868 | 1 | 21 | 0.091 | 0.0036 | n.s. |
| 1-Rec | Cyclin G1 | 0.956 | 0.053 | 0.333 | 0.967 | ||||
| P | Geminin | 0 | 0 | 0 | 0.005 | 3 | 0.045 | 0.0036 | n.s |
| 1-Rec | Geminin | 0.015 | 0.017 | 0 | 0.061 | ||||
| P | Ki67 B56 | 0.027 | 0.045 | 0 | 0.096 | 22 | 0.32 | 0.0036 | n.s. |
| 1-Rec | Ki67 B56 | 0.047 | 0.017 | 0.007 | 0.118 | ||||
| P | Ki67 Mib1 | 0 | 0.012 | 0 | 0.048 | 17 | 0.012 | 0.0036 | n.s. |
| 1-Rec | Ki67 Mib1 | 0.034 | 0.039 | 0 | 0.172 | ||||
| P | Ki67 SP6 | 0.955 | 0.054 | 0 | 1 | 13 | 1 | 0.0036 | n.s. |
| 1-Rec | Ki67 SP6 | 0.75 | 0.958 | 0.012 | 1 | ||||
| P | MCM2 | 0.021 | 0.072 | 0 | 0.118 | 13.5 | 0.52 | 0.0036 | n.s. |
| 1-Rec | MCM2 | 0.051 | 0.158 | 0 | 0.225 | ||||
| P | MCM6 | 0.5 | 0.544 | 0.174 | 1 | 20 | 0.15 | 0.0036 | n.s. |
| 1-Rec | MCM6 | 0.209 | 0.265 | 0.061 | 0.438 | ||||
| P | p15INK4b | 0.773 | 0.162 | 0.447 | 0.967 | 13 | 1 | 0.0036 | n.s |
| 1-Rec | p15INK4b | 0.884 | 0.942 | 0 | 1 | ||||
| P | p16INK4a | 0.032 | 0.045 | 0 | 0.233 | 10 | 0.69 | 0.0036 | n.s. |
| 1-Rec | p16 INK4a | 0.026 | 0.312 | 0.009 | 1 | ||||
| P | p21WAF1 | 0.818 | 0.162 | 0.264 | 1 | 32 | 0.31 | 0.0036 | n.s. |
| 1-Rec | p21WAF1 | 0.902 | 0.082 | 0.392 | 1 | ||||
Ratios of cell cycle marker positive GC nuclei in primary and first recurrent GCTB.
Bold p-values for geminin and Ki67 Mib1 indicate non-adjusted statistical significance.
Of the 3 clones for the general proliferation marker protein Ki67, both mouse monoclonals (Mib1 and B56) showed occasional positive reaction in a few GC nuclei (Figure 2A,B). Interestingly, weak to moderate Mib1 reaction generally appeared also in the cytoplasm of GCs. Unexpectedly, the rabbit monoclonal SP6 reacted in many GC nuclei, however, weaker than in the adjacent proliferating mononuclear cells (Figure 2C). Of note, strong SP6 nuclear staining in GCs was at similar frequency as with Mib1 or B56. Of the replication licensing complex proteins, MCM2 was detected also only occasionally in a few GC nuclei (Figure 2D), as opposed to MCM6 which was seen frequently, but also weaker in the large (>40 nuclei) GCs, than in the small (<10–15 nuclei) ones or in the mononuclear cells (Figure 2E). Though Mib1 positive nuclei in GCs were markedly higher (W = 17, p = 0.012) in 1-Rec than in P cases (Figure 2F), it did not reach significance after adjusting for multiple testing (p* = 0.0036). Also, neither B56 (p = 0.32) and SP6 (p = 1.0) Ki67 clones, nor MCM2 (p = 0.52) and MCM6 (p = 0.15) positive GC nuclei showed statistically different frequency between P and 1-Rec cases (see also in Table 2).
FIGURE 2
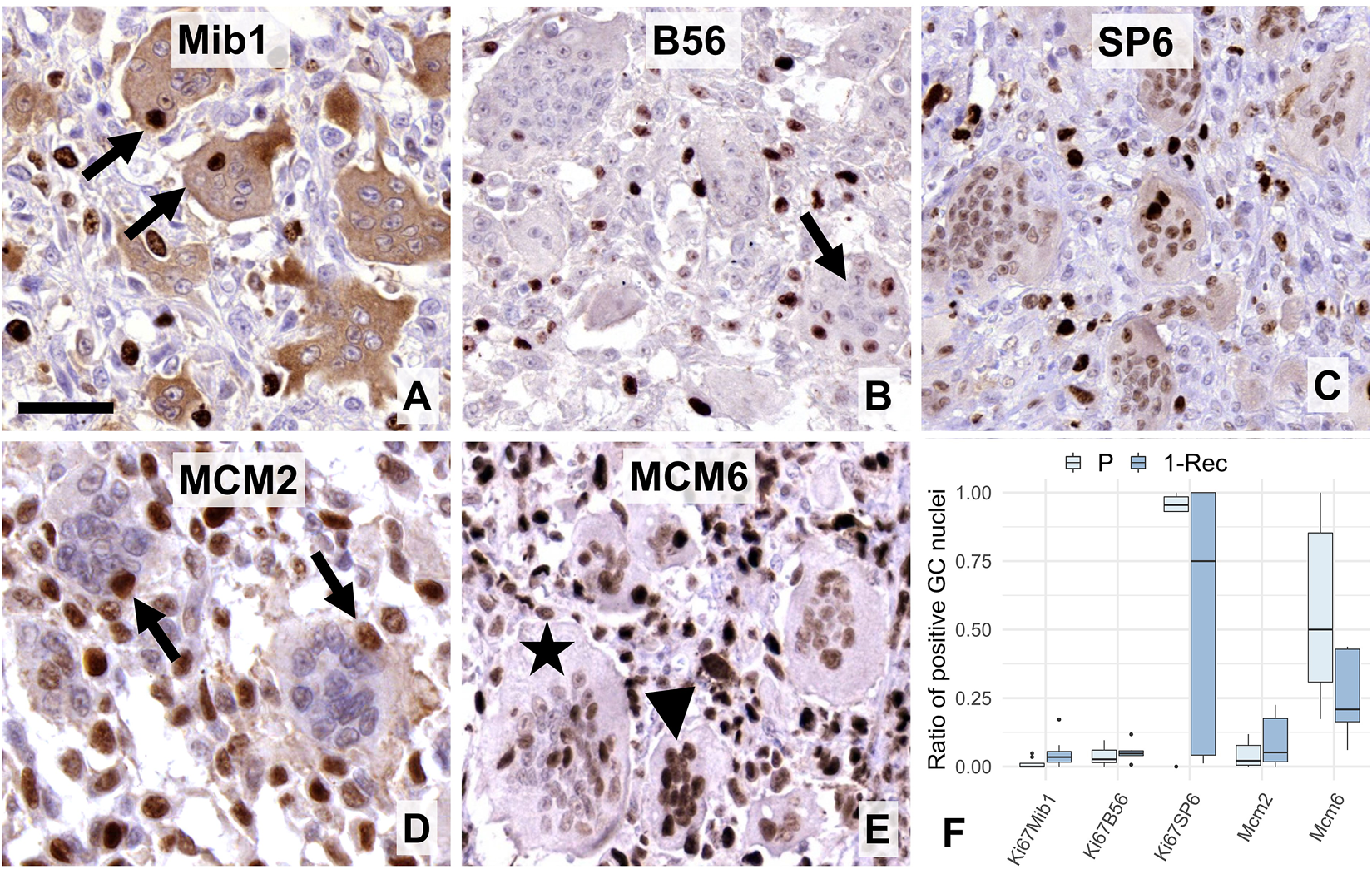
Expression of “general” proliferation markers i.e. Ki67 (A–C) and MCM-complex proteins (D–E) in multinucleated GCs. Mib1 (A) and B56 (B) antibodies showed occasional nuclear immunoreactions (arrows), while the SP6 clone resulted in usually weaker, but a widespread Ki67 positivity in GCs. Cytoplasmic Mib1 positivity in GCs was validated by negativity in several mononuclear cells. MCM2 reaction (arrows) was also rare in GCs (D), while that of MCM6 was rather frequent (E) and obviously more pronounced in smaller (arrowhead), than larger GCs (asterisk). Scale bar represents 40 μm on A, 50 μm on (B,C,E), and 30 μm on (D). Boxplot of the ratio of immunopositive GC nuclei vs. all GC nuclei (F) in primary (P) and first recurrent (1-Rec) GCTB cases.
G1/S-phase Progression Markers in Giant Cells
Concerning early G1-S phase regulation associated markers, weak to moderate cyclin dependent kinase 4/6 (CDK4/6) reactions were seen in around half of the GC nuclei (Figure 3A), while the vast majority of their nuclei showed moderate to strong reaction with the complexing partner cyclin D1 (Figure 3B). The intensity of the reactions and the rate of positive nuclei for cyclin D1 showed inverse relationship with the size and nuclear density of GCs (Figure 3C). Compared to CDK4/6, much less CDK2 positive nuclei were seen in GCs (Figure 3D). However, its complexing partner cyclin E, though it was not systematically counted, was widely detected in GC nuclei as a moderate reaction (Figure 3E), so as cyclin G1 (Figure 3F). Though average frequency of cyclin G1 in GCs showed a non-significant trend (W = 21. p = 0.091) toward P cases, CDK4 (W = 28, p = 0.72), cyclin D1 (W = 66, p = 0.25) and CDK2 (W = 72, p = 0.10) values did not differ statistically between primary and 1-Rec cases (Figure 3G).
FIGURE 3
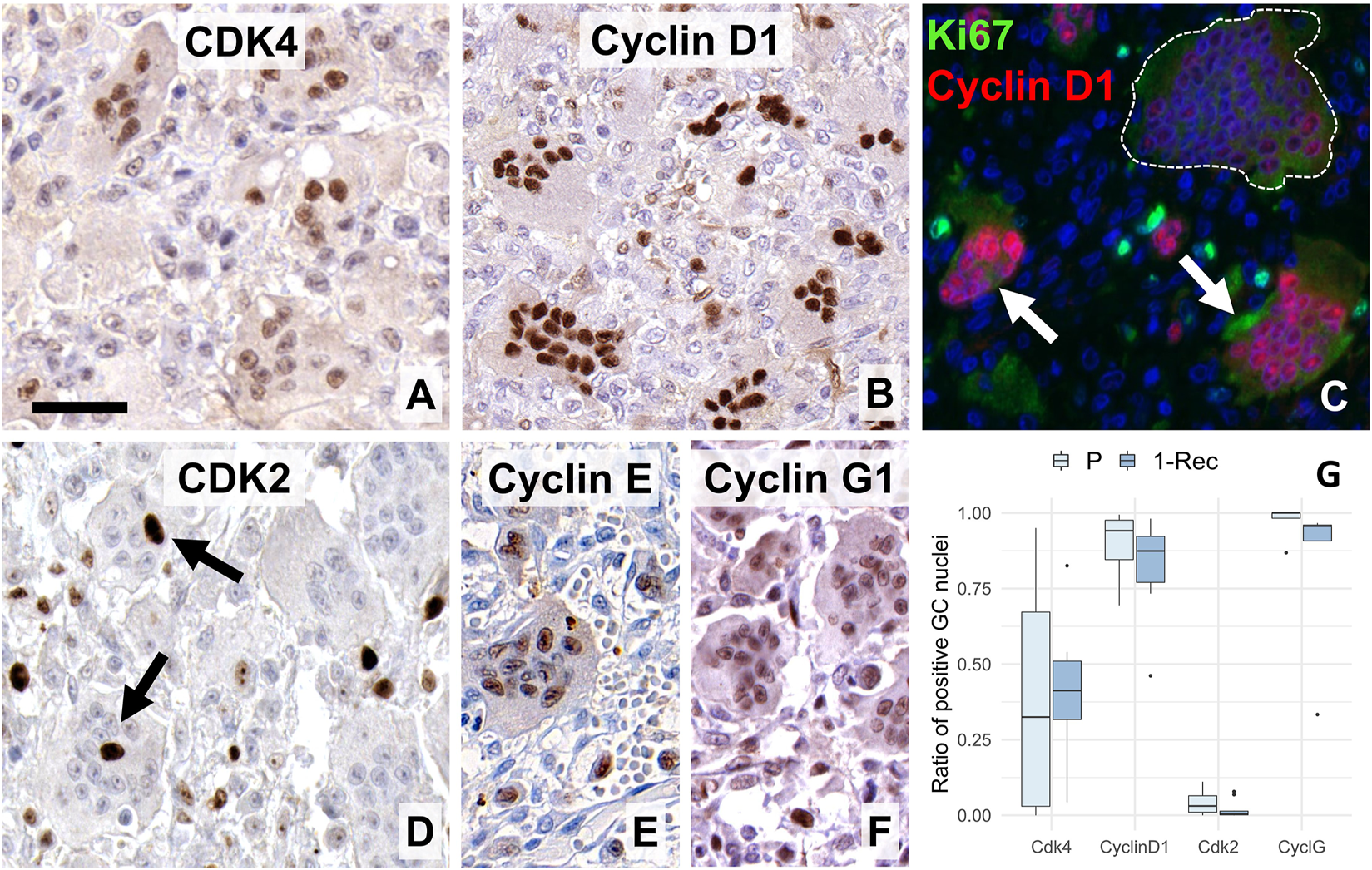
Expression of early G1-S-phase promoting cell cycle markers in giant cells (GC). Many nuclei were positive for the CDK 4/6 (A) and almost all were strongly stained for its complexing partner cyclin D1 (B). Double immunofluorescence (C) for Ki67 (Mib1, green) and cyclin D1 (SP4, red) showed strong cyclin D1 positivity in smaller GCs (white arrow) but only very faint and missing reaction in a large, aged GC (within white broken line). CDK2 (D) was rarely detected in GCs (arrows), as opposed to the widespread expression of cyclin E (E) and cyclin G1 (F). Scale bar: 40 μm for (A,C,D); and 50 μm for (B,E,F) images. Boxplots of the ratio of immunopositive GC nuclei vs. all GC nuclei (G) for some of these markers in primary (P) and first recurrence (1-Rec) GCTB.
Post G1-phase Markers and Cell Cycle Inhibitors in Giant Cells
Cyclin A, the S-G2-M transition partner of CDK2, was practically not detected (Figure 4A) in GCs, while the cell cycle repressor geminin (Figure 4B) only at very low frequency, although it appeared more often in 1-Rec cases (W = 3, p = 0.045/n.s.). Topoisomerase 2a (Figure 4C), responsible for genome organization in S-phase and chromatid segregation in mitosis, was not detected. Also, as expected from these, both the G2-M phase associated aurora kinase A and pHH3 (the latter primarily labels metaphase cells), were missing from GCs. The latter was also very rare even in the mononuclear cell fraction. Boxplots of quantitative analyses for some of these markers are shown in (Figure 4D).
FIGURE 4
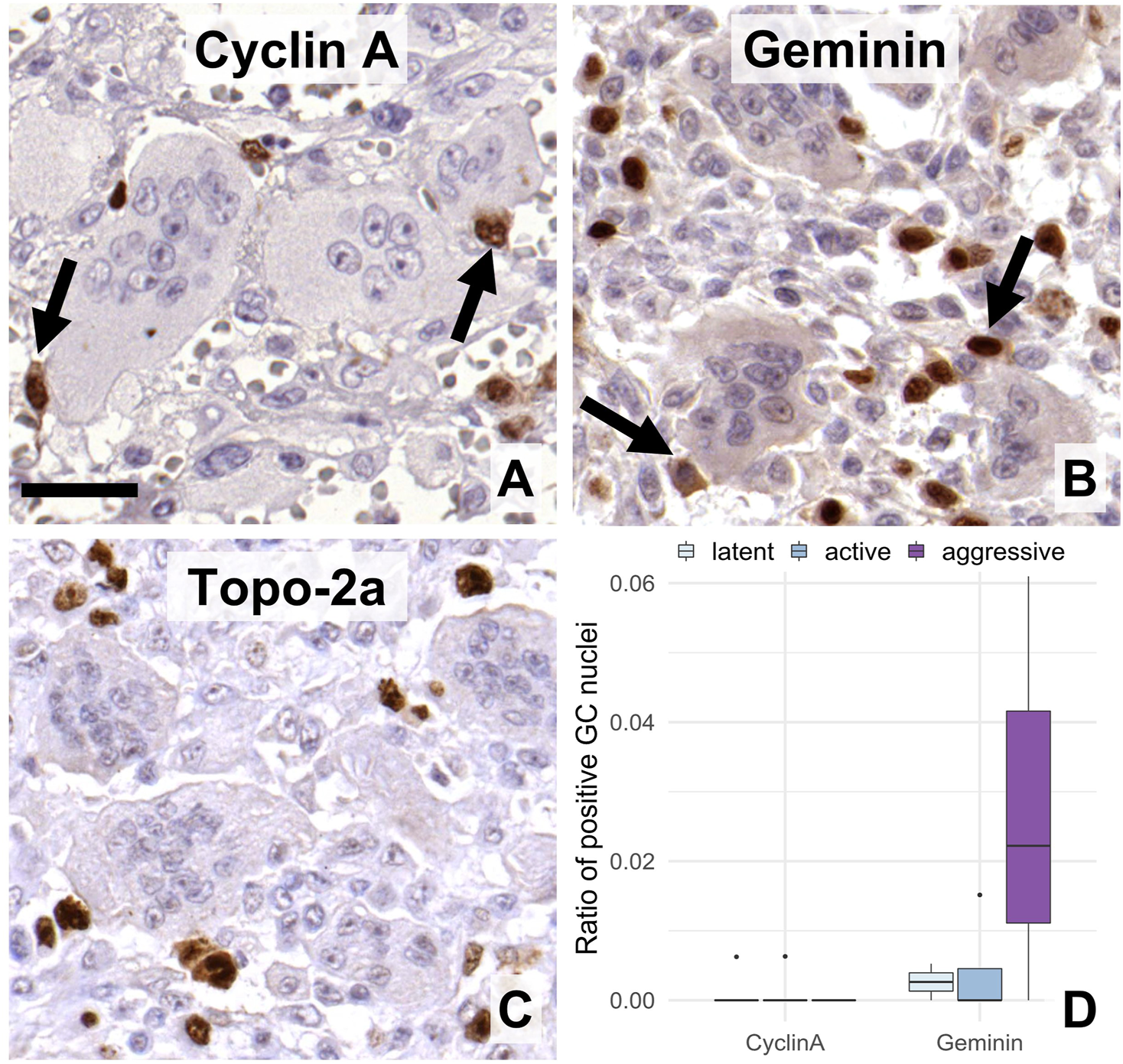
Expression of post-G1 (S-G2-M)-phase cell cycle markers in GC. Cyclin A (A), geminin (B), and topoisomerase-2a (C) were practically detected only in the mononuclear cells. Arrows show immunopositive mononuclear cells of close association with multinucleated GCs. Scale bar: 30 μm for all images. Very rare geminin positive cells were somewhat more frequent in agressive grade tumors vs. the other groups ((D), boxlot).
In line with these, all CDK inhibitors tested including p15INK4b, p16INK4a, p21WAF1 and p27KIP1 were detected widely in GC nuclei (Figures 5A–D). p16INK4a showed the least nuclear positivity but showed widespread cytoplasmic reaction, while p21WAF1 was strongly detected practically in most GC nuclei. Fitting into this pattern, the majority of GC nuclei were immunopositive over a wide range of intensities when using either the DO7 (Figure 5E) or BP53–12 antibody clones (not shown) specific for the p53; or when retinoblastoma antibody was used (Figure 5F). However, none of the systematically analyzed CDK inhibitors including p15INK4b (W = 13, p = 1.0), p16INK4a (W = 10, p = 0.69) and p21WAF1 (W = 32, p = 0.31) showed differential expression between primary and 1-Rec samples (Figures 5G,H).
FIGURE 5
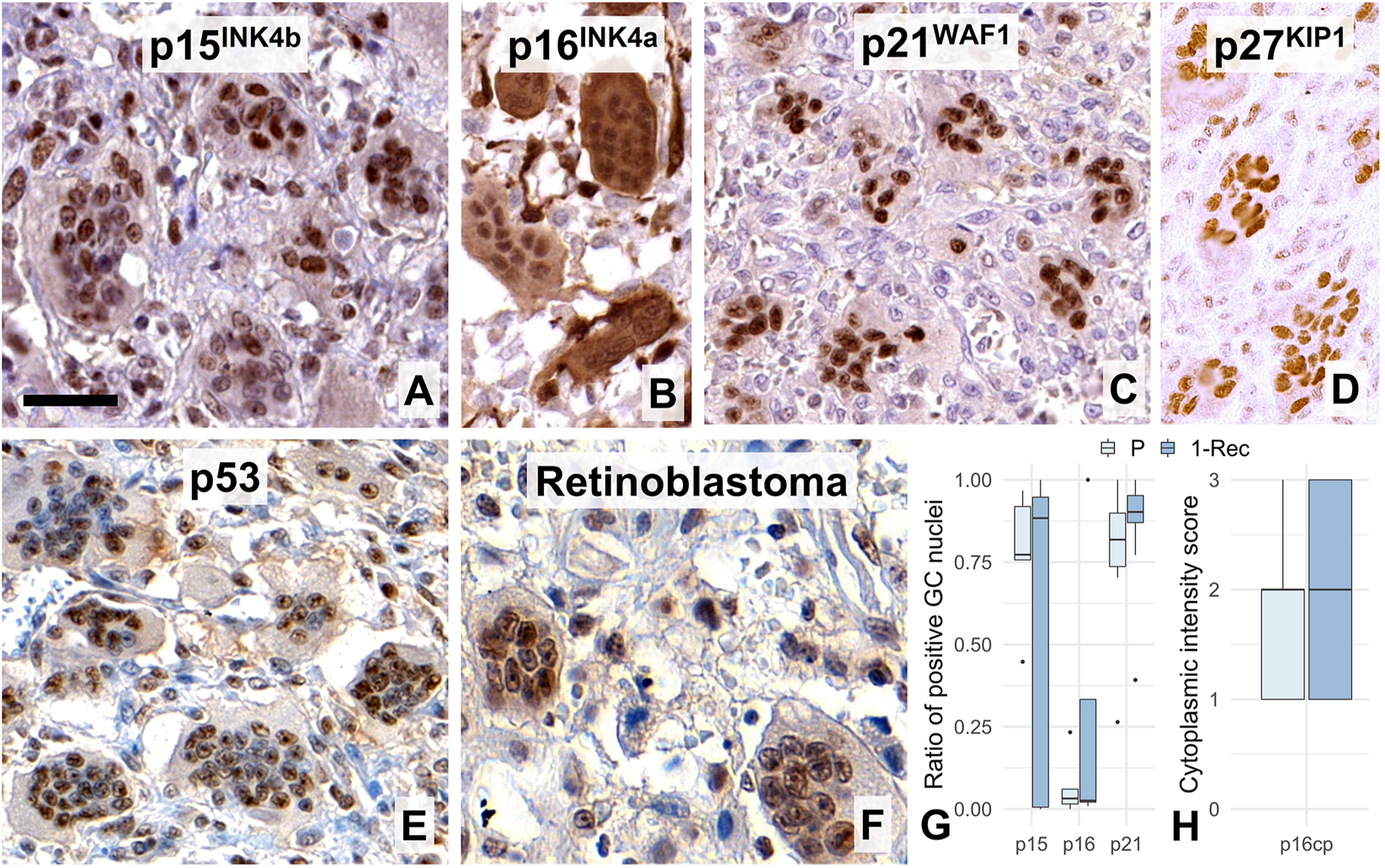
Widespread nuclear immunoreactions for the cell cycle control inhibitor markers p15INK4b(A), p16INK4a (B, where cytoplasmic reaction was frequent), p21WAF1, (C), p27KIP1(D), p53 (DO7, E), and retinoblastoma (F) proteins in multinucleated giant cell nuclei. Scale bar represents 40 μm on (A), 50 μm on (B–E), and 30 μm on (F). Boxplots of the ratio of immunopositive GC nuclei vs. all GC nuclei for some of these markers (G); and for the cytoplasmic p16INK4a reaction (H) in primary (P) and first recurrence (1-Rec) GCTBs.
Explorative Survival Analyses
Explorative univariate Cox proportional hazards analyses revealed that the increased average number of Ki67 Mib1 positive GC nuclei (HR = 1.1, 95% CI: 1–1.2, pnon-adj = 0.041) was significantly associated with shorter PFS while no other marker showed relevant associations with PFS.
Discussion
In GCTB, GCs need continuous supply of precursors to fuse, progressively form and resorb bone. They are under the influence of growth factors including receptor activator for NFκB ligand (RANKL) and macrophage colony stimulating factor (M-CSF) and their substitutes e.g. interleukins 6, 11 and 8, and TNFα; as well as VEGF, placental growth factor (PlGF), hepatocyte growth factor (HGF) and FLt-3 ligand, respectively, some of which can induce proliferation [11, 20, 21]. Earlier studies by others showed the widespread expression of cyclin D1 [5, 6, 22–24] cyclin D3 [5] and p21WAF1 [6], less p16 and scarce Ki67 [22] in GCs. Still, these data were insufficient to declare replication activity in GCs. In our present work, in addition to confirming previous observations, we revealed further cell cycle promoters, which can verify [23] an early cell cycle course in GCs. However, the effect of the generally detected cell cycle licensing MCM6, and the cell cycle promoters CDK4 and cyclin E, are likely neutralized in GCs by the upregulation CDK inhibitors p15INK4b, p16INK4a, p27KIP1 and the p53 induced p21WAF1, consistent with a cell cycle arrest at the late G1 phase. The potential emergence and links among the markers of this study during the cell cycle are drafted in Figure 6.
FIGURE 6
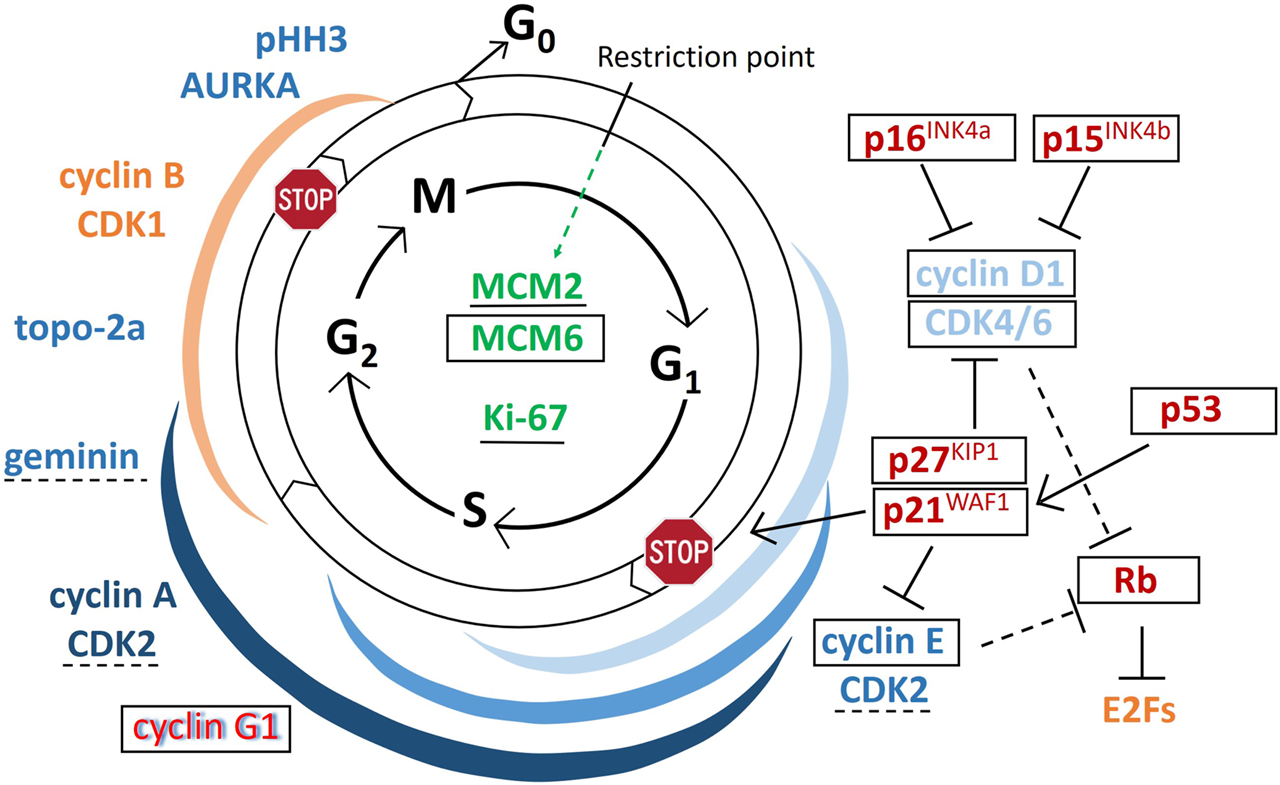
Draft on the emergence and potential role of regulatory proteins during the cell cycle. Arrows indicate activating functions while “T” signs show inhibitory functions. Markers in green can be detected throughout the cycle, those in blue support, while those in red inhibit cell cycle progression; red letters with blue shadow at cyclin G1 indicate binary functions; and the markers in yellow are important but not tested in this study. In GCTB giant cells, the markers that are framed in black were widely detected, those underlined with continuous line were occasionally found, those underlined using broken lines are found very rarely, while those which are not labeled either of these ways, were practically not detected within GCs. Colored ribbons show the rough expression duration of the matching-colored cyclin-cyclin dependent kinase complexes.
The general proliferation marker Ki67, which has been involved in heterochromatin organization, the inhibition of p21-mediated G1/S-phase checkpoint activation and in the formation of mitotic perichromosomal protein sheet, can be detected throughout the replication cycle [25]. So far, only one study reported a weak Ki67 Mib1 staining in <5% of GCs in only 3 out of 29 (10%) GCTB samples. Of the 3 Ki67 clones we investigated in GCs, both Mib1 and B56 showed a low ratio of nuclear positivity (<10%) while SP6 demonstrated a wide range with >75% median. The specificity of the moderate intensity SP6 reaction in GCs was validated by the clear negativity of some adjacent mononuclear cells. Though Ki67 expression peaks at G2/M-phases, the protein is only gradually eliminated from newly divided cells, therefore, its amount may also reflect the time the cell spent in quiescence [26]. Accordingly, the markedly higher ratio of positive GC nuclei in 1-Rec vs. primary cases, and its association with shorter PFS in our explorative survival analysis may also indicate accelerated dynamics of GC formation in the advanced cases. The cytoplasmic Mib1 staining in GCs shown also by others [27] is likely to be related to its metabolic elimination by the ubiquitin proteasome system [28]. The differential occurrence of the Ki67 clones can be partly related to the slower degradation of the epitope region recognized by SP6 compared to the others. However, further signs in this study suggested that this may also be linked with an initial replication activity.
Members of the MCM2-7 helicase complex, which can also be detected all over the cycle, unwind double-stranded DNA and allow the controlled licensing of DNA for duplication [16]. However, while MCM6 complexed with MCM4 and MCM7 is involved in relaxing DNA to single strands, the MCM2 subunit (MCM3, and MCM5 too), has potential inhibitory role on this function [29]. Our widespread detection of MCM6 with moderate intensity but only occasional occurrence of MCM2 in GC nuclei, may reflect the initiation of DNA unwinding in GCs.
Cyclin-dependent serine/threonine kinases complex with their regulatory subunit cyclins to phosphorylate retinoblastoma and promote the transition of different phases during cell cycle progression [30]. Here, we revealed the widespread emergence not only of the previously detected cyclin D1 [5, 6], but also its partner CDK4 in the earliest G1-S phase promoter complex. This complex not only can inactivate retinoblastoma but also support the activation of the next G1-S-phase promoter cyclin E-CDK2 complex through reducing mitochondrial metabolism to prevent cyclin E degradation [31]. In agreement with this, we regularly detected cyclin E in GCs, however, only very rarely found its complexing partner CDK2 in GC nuclei, indicating a late G1-phase arrested cell cycle. At the same time, cyclin D1-CDK4/6 complex can also support cell growth e.g. through mTORC1 activation [32], and high cyclin D1 levels can assist in metabolic substrate utilization toward mitochondrial amino acid production [31], which propose a role for cyclin D1 also in GC growth and differentiation. Furthermore, cyclin D1 overexpression in GCs may also be required for multinucleation [5], as it enhanced the number of nuclei e.g. in cardiomyocytes [33] and was detected primarily in giant trophoblasts rather than in diploid ones [34].
In agreement with earlier studies, in GCs we also revealed the widespread expression of p21WAF1, the universal CDK inhibitor [35]. However, despite p21WAF1 is considered to block cyclin D1-CDK4/6 complex and CDK2 activities, paradoxically it may also be important for CDK4/6 complex assembly [36] and for the nuclear export of cyclin D1 [37, 38]. These might explain our frequent detection of CDK4 but not the CDK2, and the high levels of the CDK4 partner cyclin D1 in GC nuclei. P21WAF1 is an important effector of the cell cycle control functions of p53 [39]. Before us, p53 expression had been noticed only incidentally in one publication [40]. Here, we detected p53 in the majority of GC nuclei using 2 different antibody clones (DO7 and BP53–12 respectively) to give, for the first time, clear evidence that GCs upregulate this important “guardian” of the genome. Cytoskeletal stress, which may occur during multinucleation in GCs, can induce p53 upregulation and activate p21WAF1 for blocking S-phase entry at G1-checkpoint [39]. The p53-dependent G1 arrest of multinucleated tetraploid cells has been previously described [41] and p53 activity with the contribution of p21WAF1 can also drive this process even further to result in cellular senescence [42]. We also noticed the general upregulation of p27KIP1 in GCs, which in normal cells is primarily linked to controlling the cell cycle through inhibiting both cyclin E-CDK2 and cyclin D1-CDK4/6 activities [43]. Both p21WAF1 and p27KIP1 play essential roles also in GC functions as shown by the osteopetrotic phenotype in mice with deleted genes encoding these proteins [44]. In GCTB, arresting of the potential cell cycle activities and inducing senescence are required for GC maturation involving the production of key proteases cathepsin K and MMP-9 for pathological bone resorption [45].
As a p53 target, cyclin G1 has been involved in both supporting cell cycle arrest and in driving the S-G2-M-transition [17]. As a supposed oncogene cyclin G1 may activate the MDM2 oncoprotein by recruiting Ser/Thr protein phosphatase 2A (PP2A), which dephosphorylates MDM2 to inhibit and degrade p53 [17, 18, 46]. We detected cyclin G1 in most GC nuclei with a non-significant trend toward higher values in primary vs. 1-Rec GCTB cases. This may rather support its role in cell cycle arrest than as a promoter of replication in GCs which are reactive, non-malignant cells. However, it is also possible, that cyclin G1 upregulation is a rebound effect to control p53 overexpression and prevent apoptosis induction [17, 18, 46]. The permanently elevated cyclin D1 levels in functionally active GCs, but its disappearance from oversized (>40 nuclei), aged GCs may also be consistent with the protective function of cyclin D1 [17, 46].
Besides p21WAF1 and p27KIP1, the upregulation of other CDK inhibitors, particularly targeting CDK4 and CDK6 proteins including p15INK4b and p16INK4a, further support the cell cycle related activity in GCs [47]. Both p15INK4b and p16INK4a are also likely to contribute to G1-phase cell cycle arrest [48, 49], while p16INK4a may also be linked to cellular senescence induction supporting either the full functional differentiation or the aged-cell decay in GCs [50]. Cell cycle arrest by CDK inhibitors was also reflected by retinoblastoma upregulation in GCs. In line with these findings, practically none of the S-G2-M phase markers including cyclin A, the later complexing partner of CDK2, the cell cycle repressor geminin, the G2-M phase transition associated cyclin B, or the M-phase related AURKA and pHH3 were detected within GCs [16]. This pattern is consistent with the full functional maturation of GCs. The S-G2 phase marker positive mononuclear cells which had no discernible cytoplasmic boarders with GCs, were likely to be monocytes close to or within the fusion process (Figures 4A,B). In support of this, a subpopulation of CD14+/CD33 + monocytes was shown to proliferate in response to M-CSF and form pre-osteoclasts when primed with RANKL [51]. Then pre-osteoclasts loose CD33 expression and fuse with more monocytic cells [52] and the proliferating monocytes form significantly more GCs than the rest of the monocytic pool [51].
Despite silenced, the early signs of replication proved here, may reflect the generation dynamics and age-related activity of GCs. In vitro data confirm an inverse correlation between GC size and resorption activity at mild acidic conditions [53]. This is in line with data showing elevated recurrence potential in GCTB cases, which dominantly contain GCs of <15 nuclei compared to those carrying larger ones [18]. In the present study, the average size and nuclear density of GCs also revealed an inverse trend with the clinicoradiological grade of GCTB. Furthermore, the cyclin D1 and MCM6 reactions in GCs were obviously stronger and more frequent in small sized (∼<15 nuclei) than in larger GCs, particularly when nuclear numbers were ∼>40. These are also in agreement with our earlier finding of the significantly lower average size of GCs in recurrent GCTB cases where the growth related EGFR protein level was elevated in the mononuclear stromal cells, the major drivers of GC formation and activity [13]. All these support the view that smaller sized GCs are the younger, dynamically forming populations, which may show more signs of early replication than the aged, functionally less active oversized GCs (∼>40 nuclei).
Dissecting the replication cycle into major cells fractions through the immunohistochemical detection of nuclear proteins which regulate or control different phases of replication, may allow accurate assessment of cell proliferation dynamics in situ. Using a marker set, selected carefully from the list we described here, can serve cell cycle analysis for diagnostic, prognostic and predictive purposes in any pathological process particularly in cancer.
Conclusion
Though multinucleated GCs in GCTB are thought to be of reactive phenotype formed by fusion of cells of the monocyte macrophage lineage, they had been occasionally shown to express cell proliferation related markers. By using a comprehensive marker set, here we revealed, for the first time in GCs, the general upregulation of cell cycle promoting markers MCM6, CDK4 and cyclin E, indicating primary DNA unwinding and G1-S-phase promoting activities. We also confirmed the earlier published widespread expression of cyclin D1, which all, unequivocally demonstrated an early replication activity in GCs. This, however, was silenced by the widespread expression of CDK inhibitors p15INK4b, p16INK4a, p27KIP1 and p53 induced p21WAF1 resulting in cell cycle arrest at the G1 checkpoint, confirmed by the missing production of post-G1-phase faction markers. The complex interplay among these elements under the influence of external growth and differentiation factors is required for the functional maturation and bone resorbing activity of GCs in GCTB.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the Institute of Rizzoli (IOR), Bologna, Italy (ethical approval no.: IOR 13351/5-28-2008), and Semmelweis University, Budapest, Hungary (ethical approval no.: 87/2007). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
Conceived and designed the experiments: TK and MM. Performed the experiments: MM, PB, and PP. Analyzed the data: MM, TK, HW, and PP. Performed statistical analyses: MM; Contributed reagents/materials/analysis tools: PP, ZS, MS, RF, CG, and TK. Wrote the paper: MM and TK. Diagnosed GCTB cases and advised the study: ZS, MS, and RF. Supervised the study: TK. All authors critically reviewed the manuscript and approved the final version.
Funding
This study was performed within the framework of EuroBoNet, an EU FP6 funded Network of Excellence Program (LSHC-CT-2006-018814). The founders had no influence on study design, data collection, analysis and decision to publish, or preparation of the manuscript.
Conflict of interest
MM reports non-related funding from the German Ministry for Education and Research (MI-I 01ZZ1801E) and non-related consultancy for Siemens Healthineers and SmartReporting GmbH. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Chambers TJ Multinucleate Giant Cells. J Pathol (1978) 126(3):125–48. 10.1002/path.1711260302
2.
Ariizumi T Ogose A Kawashima H Hotta T Umezu H Endo N Multinucleation Followed by an Acytokinetic Cell Division in Myxofibrosarcoma with Giant Cell Proliferation. J Exp Clin Cancer Res (2009) 28:44. 10.1186/1756-9966-28-44
3.
Vignery A Macrophage Fusion. J Exp Med (2005) 202(3):337–40. 10.1084/jem.20051123
4.
Maros ME Schnaidt S Balla P Kelemen Z Sapi Z Szendroi M et al In situ cell Cycle Analysis in Giant Cell Tumor of Bone Reveals Patients with Elevated Risk of Reduced Progression-free Survival. Bone (2019) 127:188–98. 10.1016/j.bone.2019.06.022
5.
Kauzman A Li SQ Bradley G Bell RS Wunder JS Kandel R Cyclin Alterations in Giant Cell Tumor of Bone. Mod Pathol (2003) 16(3):210–8. 10.1097/01.MP.0000057235.65327.40
6.
Kandel R Li SQ Bell R Wunder J Ferguson P Kauzman A et al Cyclin D1 and P21 Is Elevated in the Giant Cells of Giant Cell Tumors. J Orthop Res (2006) 24(3):428–37. 10.1002/jor.20036
7.
Maggiani F Forsyth R Hogendoorn PCW Krenacs T Athanasou NA The Immunophenotype of Osteoclasts and Macrophage Polykaryons. J Clin Pathol (2011) 64(8):701–5. 10.1136/jcp.2011.090852
8.
Behjati S Tarpey PS Presneau N Scheipl S Pillay N Van Loo P et al Distinct H3F3A and H3F3B Driver Mutations Define Chondroblastoma and Giant Cell Tumor of Bone. Nat Genet (2013) 45(12):1479–82. 10.1038/ng.2814
9.
Szendrői M Giant-cell Tumour of Bone (GCT). In: BentleyG, editor. European Surgical Orthopaedics and Traumatology. Berlin, Germany: Springer (2014). p. 4037–54.
10.
Cowan RW Singh G Giant Cell Tumor of Bone: A Basic Science Perspective. Bone (2013) 52(1):238–46. 10.1016/j.bone.2012.10.002
11.
Knowles HJ Athanasou NA Acute Hypoxia and Osteoclast Activity: a Balance between Enhanced Resorption and Increased Apoptosis. J Pathol (2009) 218(2):256–64. 10.1002/path.2534
12.
Knowles HJ Athanasou NA Canonical and Non-canonical Pathways of Osteoclast Formation. Histol Histopathol (2009) 24:337. 10.14670/HH-24.337
13.
Balla P Moskovszky L Sapi Z Forsyth R Knowles H Athanasou NA et al Epidermal Growth Factor Receptor Signalling Contributes to Osteoblastic Stromal Cell Proliferation, Osteoclastogenesis and Disease Progression in Giant Cell Tumour of Bone. Histopathology (2011) 59(3):376–89. 10.1111/j.1365-2559.2011.03948.x
14.
Balla P Maros ME Barna G Antal I Papp G Sapi Z et al Prognostic Impact of Reduced Connexin43 Expression and Gap Junction Coupling of Neoplastic Stromal Cells in Giant Cell Tumor of Bone. PLoS One (2015) 10(5):e0125316. 10.1371/journal.pone.0125316
15.
Satyanarayana A Kaldis P Mammalian Cell-Cycle Regulation: Several Cdks, Numerous Cyclins and Diverse Compensatory Mechanisms. Oncogene (2009) 28(33):2925–39. 10.1038/onc.2009.170
16.
Williams GH Stoeber K The Cell Cycle and Cancer. J Pathol (2012) 226(2):352–64. 10.1002/path.3022
17.
Kimura SH Nojima H Cyclin G1 Associates with MDM2 and Regulates Accumulation and Degradation of P53 Protein. Genes Cells (2002) 7(8):869–80. 10.1046/j.1365-2443.2002.00564.x
18.
Gordon E Ravicz J Liu S Chawla S Hall F Cell Cycle Checkpoint Control: The Cyclin G1/Mdm2/p53 axis Emerges as a Strategic Target for Broad-Spectrum Cancer Gene Therapy - A Review of Molecular Mechanisms for Oncologists. Mol Clin Onc (2018) 9(2):115–34. 10.3892/mco.2018.1657
19.
Wenz H Maros ME Meyer M Förster A Haubenreisser H Kurth S et al Image Quality of 3rd Generation Spiral Cranial Dual-Source CT in Combination with an Advanced Model Iterative Reconstruction Technique: A Prospective Intra-individual Comparison Study to Standard Sequential Cranial CT Using Identical Radiation Dose. PLoS One (2015) 10(8):e0136054. 10.1371/journal.pone.0136054
20.
Knowles H Athanasou N Hypoxia-inducible Factor Is Expressed in Giant Cell Tumour of Bone and Mediates Paracrine Effects of Hypoxia on Monocyte-Osteoclast Differentiation via Induction of VEGF. J Pathol (2008) 215(1):56–66. 10.1002/path.2319
21.
Knowles HJ Athanasou NA Canonical and Non-canonical Pathways of Osteoclast Formation. Histol Histopathol (2009) 24(3):337–46. 10.14670/HH-24.337
22.
Choi JW Lee JH Kim YS Kim YS Frequent Upregulation of Cyclin D1 and p16INK4a Expression with Low Ki-67 Scores in Multinucleated Giant Cells. Pathobiology (2011) 78(4):233–7. 10.1159/000327359
23.
Matsubayashi S Nakashima M Kumagai K Egashira M Naruke Y Kondo H et al Immunohistochemical Analyses of β-catenin and Cyclin D1 Expression in Giant Cell Tumor of Bone (GCTB): A Possible Role of Wnt Pathway in GCTB Tumorigenesis. Pathol - Res Pract (2009) 205(9):626–33. 10.1016/j.prp.2009.02.011
24.
Lujic N Sopta J Kovacevic R Stevanovic V Davidovic R Recurrence of Giant Cell Tumour of Bone: Role of P53, Cyclin D1, β-catenin and Ki67. Int Orthopaedics (Sicot) (2016) 40(11):2393–9. 10.1007/s00264-016-3292-2
25.
Sun X Kaufman PD Ki-67: More Than a Proliferation Marker. Chromosoma (2018) 127(2):175–86. 10.1007/s00412-018-0659-8
26.
Sales Gil R Vagnarelli P Ki-67: More Hidden behind a 'Classic Proliferation Marker'. Trends Biochem Sci (2018) 43(10):747–8. 10.1016/j.tibs.2018.08.004
27.
Ismail FW Shamsudin AM Wan Z Daud SM Samarendra MS Ki-67 Immuno-Histochemistry Index in Stage III Giant Cell Tumor of the Bone. J Exp Clin Cancer Res (2010) 29:25. 10.1186/1756-9966-29-25
28.
Miller I Min M Yang C Tian C Gookin S Carter D et al Ki67 Is a Graded Rather Than a Binary Marker of Proliferation versus Quiescence. Cell Rep (2018) 24(5):1105–12.e5. 10.1016/j.celrep.2018.06.110
29.
Vijayraghavan S Schwacha A The Eukaryotic Mcm2-7 Replicative Helicase. Subcell Biochem (2012) 62:113–34. 10.1007/978-94-007-4572-8_7
30.
Malumbres M Cyclin-dependent Kinases. Genome Biol (2014) 15(6):122. 10.1186/gb4184
31.
Pestell RG New Roles of Cyclin D1. Am J Pathol (2013) 183(1):3–9. 10.1016/j.ajpath.2013.03.001
32.
Romero-Pozuelo J Figlia G Kaya O Martin-Villalba A Teleman AA Cdk4 and Cdk6 Couple the Cell-Cycle Machinery to Cell Growth via mTORC1. Cell Rep (2020) 31(2):107504. 10.1016/j.celrep.2020.03.068
33.
Soonpaa MH Koh GY Pajak L Jing S Wang H Franklin MT et al Cyclin D1 Overexpression Promotes Cardiomyocyte DNA Synthesis and Multinucleation in Transgenic Mice. J Clin Invest (1997) 99(11):2644–54. 10.1172/JCI119453
34.
Palazon LS Davies TJ Gardner RL Translational Inhibition of Cyclin B1 and Appearance of Cyclin D1 Very Early in the Differentiation of Mouse Trophoblast Giant Cells. Mol Hum Reprod (1998) 4(11):1013–20. 10.1093/molehr/4.11.1013
35.
Georgakilas AG Martin OA Bonner WM p21: A Two-Faced Genome Guardian. Trends Mol Med (2017) 23(4):310–9. 10.1016/j.molmed.2017.02.001
36.
Cheng M Olivier P Diehl JA Fero M Roussel MF Roberts JM et al The p21Cip1 and p27Kip1 CDK `inhibitors' Are Essential Activators of Cyclin D-dependent Kinases in Murine Fibroblasts. EMBO J (1999) 18(6):1571–83. 10.1093/emboj/18.6.1571
37.
Alt JR Cleveland JL Hannink M Diehl JA Phosphorylation-dependent Regulation of Cyclin D1 Nuclear Export and Cyclin D1-dependent Cellular Transformation. Genes Dev (2000) 14(24):3102–14. 10.1101/gad.854900
38.
Alt JR Gladden AB Diehl JA p21Cip1 Promotes Cyclin D1 Nuclear Accumulation via Direct Inhibition of Nuclear Export. J Biol Chem (2002) 277(10):8517–23. 10.1074/jbc.M108867200
39.
Hafner A Bulyk ML Jambhekar A Lahav G The Multiple Mechanisms that Regulate P53 Activity and Cell Fate. Nat Rev Mol Cell Biol (2019) 20(4):199–210. 10.1038/s41580-019-0110-x
40.
Yalcinkaya U Ugras N Kabul S Ocakoglu G Bilgen MS Prognostic Value of P53 Protein Expression in Giant Cell Tumor of Bone. pjp (2015) 4(4):389–96. 10.5114/pjp.2015.57252
41.
Ganem NJ Pellman D Limiting the Proliferation of Polyploid Cells. Cell (2007) 131(3):437–40. 10.1016/j.cell.2007.10.024
42.
Rieder CL Maiato H Stuck in Division or Passing through. Dev Cell (2004) 7(5):637–51. 10.1016/j.devcel.2004.09.002
43.
Wander SA Zhao D Slingerland JM p27: A Barometer of Signaling Deregulation and Potential Predictor of Response to Targeted Therapies: Figure 1. Clin Cancer Res (2011) 17(1):12–8. 10.1158/1078-0432.Ccr-10-0752
44.
Sankar U Patel K Rosol TJ Ostrowski MC RANKL Coordinates Cell Cycle Withdrawal and Differentiation in Osteoclasts Through the Cyclin-Dependent Kinase Inhibitors p27KIP1and p21CIP1. J Bone Miner Res (2004) 19(8):1339–48. 10.1359/JBMR.040321
45.
Lindeman JHN Hanemaaijer R Mulder A Dijkstra PDS Szuhai K Bromme D et al Cathepsin K Is the Principal Protease in Giant Cell Tumor of Bone. Am J Pathol (2004) 165(2):593–600. 10.1016/S0002-9440(10)63323-8
46.
Okamoto K Li H Jensen MR Zhang T Taya Y Thorgeirsson SS et al Cyclin G Recruits PP2A to Dephosphorylate Mdm2. Mol Cell (2002) 9(4):761–71. 10.1016/s1097-2765(02)00504-x
47.
Sherr CJ Roberts JM CDK Inhibitors: Positive and Negative Regulators of G1-phase Progression. Genes Dev (1999) 13(12):1501–12. 10.1101/gad.13.12.1501
48.
Carnero A Hudson JD Hannon GJ Beach DH Loss-of-function Genetics in Mammalian Cells: the P53 Tumor Suppressor Model. Nucleic Acids Res (2000) 28(11):2234–41. 10.1093/nar/28.11.2234
49.
Hudson JD Shoaibi MA Maestro R Carnero A Hannon GJ Beach DH A Proinflammatory Cytokine Inhibits P53 Tumor Suppressor Activity. J Exp Med (1999) 190(10):1375–82. 10.1084/jem.190.10.1375
50.
Dimri GP Hara E Campisi J Regulation of Two E2F-Related Genes in Presenescent and Senescent Human Fibroblasts. J Biol Chem (1994) 269(23):16180–6. 10.1016/s0021-9258(17)33990-x
51.
Lari R Kitchener PD Hamilton JA The Proliferative Human Monocyte Subpopulation Contains Osteoclast Precursors. Arthritis Res Ther (2009) 11(1):R23. 10.1186/ar2616
52.
Forsyth RG De Boeck G Baelde JJ Taminiau AH Uyttendaele D Roels H et al CD33+ CD14− Phenotype Is Characteristic of Multinuclear Osteoclast-like Cells in Giant Cell Tumor of Bone. J Bone Mineral Res (2009) 24(1):70–7. 10.1359/jbmr.080905
53.
Orriss IR Arnett TR Rodent Osteoclast Cultures. Methods Mol Biol (2012) 816:103–17. 10.1007/978-1-61779-415-5_8
Summary
Keywords
giant cell tumor of bone, giant cells, Ki-67, cyclin D1, p53, p21 (CDKN1A) WAF1, cyclin G1
Citation
Maros ME, Balla P, Micsik T, Sapi Z, Szendroi M, Wenz H, Groden C, Forsyth RG, Picci P and Krenacs T (2021) Cell Cycle Regulatory Protein Expression in Multinucleated Giant Cells of Giant Cell Tumor of Bone: do They Proliferate?. Pathol. Oncol. Res. 27:643146. doi: 10.3389/pore.2021.643146
Received
17 December 2020
Accepted
01 April 2021
Published
03 May 2021
Volume
27 - 2021
Edited by
Anna Sebestyén, Semmelweis University, Hungary
Updates
Copyright
© 2021 Maros, Balla, Micsik, Sapi, Szendroi, Wenz, Groden, Forsyth, Picci and Krenacs.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tibor Krenacs, krenacst@gmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.