- 1Division of Diagnostic Pathology, Kikuna Memorial Hospital, Yokohama, Japan
- 2Division of Pathology, Shizuoka Cancer Center, Shizuoka, Japan
- 3Department of Human Pathology, Juntendo University School of Medicine, Tokyo, Japan
- 4Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan
Recent evidence suggests that somatic mutations in ERBB2 activate ERBB2 signaling. These mutations occur at a frequency of approximately 3% in breast cancer (BC). ERBB2 mutations indicate poor prognosis as they are associated with recurrence and metastasis. This study aimed to evaluate the clinicopathological features, immune infiltration levels, tumor mutational burden (TMB), and tumor-infiltrating lymphocytes (TILs) in ERBB2-mutated breast cancer (ERBB2-mutated BC) using a bioinformatic approach and publicly available datasets (i.e., TCGA-BRCA and TIMER2.0). ERBB2-mutated BCs were associated with a high histological grade. ERBB2-mutated BCs comprised invasive breast carcinoma of no special type (21/35, 60%), classic invasive lobular carcinoma (12/35, 34.3%), and pleomorphic invasive lobular carcinoma (2/35, 5.7%). A Kaplan-Meier survival curve demonstrated that ERBB2-mutated BC was associated with a significantly worse prognosis compared to ERBB2 non-mutated BC (p < 0.01). Furthermore, 40% (14/35) of the patients with ERBB2-mutated BC harbored CDH1 mutations. Mutations at L755 and V777 accounted for 30.5% of these mutations in ERBB2-mutated BC, suggesting that these sites are mutational hot spots in BC, particularly in invasive lobular carcinoma. Of the ERBB2-mutated BCs, 8.6% were classified as TIL-high, whereas 77.1% were TILs-low; TMB significantly correlated with TILs (p < 0.05). CD8+ T cell infiltration levels were significantly higher in ERBB2 non-mutated BC. Among ERBB2-mutated BCs, 22.9% were classified as TMB-high, which was significantly higher than the rate in the ERBB2 non-mutated BC (p < 0.01). These findings provide evidence for a link between ERBB2 mutations and high TMB in BC.
Introduction
It has been recently reported that in rare cases, somatic mutations in ERBB2 can activate ERBB2 signaling [1]. In vitro analyses have demonstrated that some ERBB2 mutations are oncogenic and promote cancer cell growth, invasion, and survival [1, 2]. A systematic review revealed that the frequency of ERBB2 mutations in breast cancer (BC) is relatively low (2.7%) [3]. ERBB2 mutations are more common in invasive lobular carcinoma (ILC) than in invasive breast carcinoma of no special type (IBC-NST) [4]. ILC is classified into two subtypes, classic ILC (c-ILC) and pleomorphic ILC (p-ILC). CDH1 is located on chromosome 16q22.1 and encodes E-cadherin [5], a transmembrane glycoprotein expressed in epithelial tissues and contributes to calcium-dependent cell-to-cell adhesion [6]. Patients with relapsed CDH1-mutated ILC exhibit a higher frequency of ERBB2 somatic mutations than those with non-CDH1-mutated BC [7]. ERBB2 mutations indicate a significantly poor prognosis, regardless of the histological type [8, 9] as they are associated with relapse and bone metastasis [7, 10]. Previous studies have demonstrated that the ERBB2 mutation is frequently associated with ILC [7, 10–14]. The CDH1 mutation is a major gene mutation in ILC, but only a few studies have focused on histopathological images and mutations of ERBB2 and CDH1 [12, 14]. Moreover, the number of ERBB2-mutated BC was relatively fewer, 5 cases and 18 cases, respectively. Therefore, the relationship between the histological type and genetic alteration of ERBB2 and CDH1 has not been completely elucidated. In this study, we investigated the clinicopathological characteristics and the frequency of co-occurrence of the CDH1 mutation and tumour-infiltrating lymphocytes (TILs) in 35 ERBB2-mutated BCs via a public database (TCGA-BRCA). Furthermore, we compared the tumor mutational burden (TMB) as well as CD8+ T cell, CD4+ T cell, and Treg immune infiltration levels between ERBB2-mutated BC and ERBB2 non-mutated BC via in silico analysis.
Materials and Methods
Data Collection
Data on ERBB2 and CDH1 somatic mutations were obtained from UCSC Xena (http://xena.ucsc.edu/). Genomic Data Commons (GDC) TCGA Breast Cancer (BRCA), comprising 986 BC samples, was used to obtain mutation data. Clinicopathological information (age of onset, ethnicity, sex, histology, and subtype) and genomic information from TCGA-BRCA were obtained using the GDC Data Portal (https://portal.gdc.cancer.gov) (Supplementary Table S1). The ERBB2 mutation type, amino acid change, and mutation site were identified. The ERBB2 mutation site was classified as follows: Receptor-L domain, Furin-like cysteine-rich domain, kinase domain, interdomain region, or C-terminal region. The pathological significance of each ERBB2 or CDH1 mutation was assessed using COSMIC FATHMM, Ensembl Variant Effect Predictor (VEP), SHIFT, and PolyPhen (Supplementary Table S2). Statistical analyses were performed using R software, version 4.0.3.
Clinicopathological Features
Clinicopathological features, including age, sex, ethnicity, and intrinsic subtype data were obtained from the GDC Data Portal. Intrinsic subtypes, as determined using multi-gene assay PAM 50, were classified into the following five types: Luminal A (LumA), Luminal B (LumB), Her-2, Basal, and normal. However, there were cases in which subtypes were not assigned and others with two subtypes; these were excluded. Finally, there were 32 cases in ERBB2-mutated BC and 859 cases in ERBB2 non-mutated BC. The prognosis of ERBB2 mutated BC was compared with that of ERBB2 non-mutated BC and analyzed on the Kaplan–Meier survival curve using UCSC Xena (http://xena.ucsc.edu/).
Pathological Review
Samples with ERBB2 mutations (n = 35) were reassessed. Whole slide images of diagnostic sections in ERBB2-mutated cases (n = 35) were downloaded from the GDC Data Portal and analyzed using Aperio ImageScope (Sausalito, CA, United States). The histological types were classified according to the 5th edition of the World Health Organization classification system [15]. The histological grade was evaluated in accordance with the Elston and Ellis modification of the Nottingham grading system [16]. All samples were reviewed by two pathologists (S.U. and T.S.). Interobserver differences were resolved through re-evaluation and discussion to reach consensus. Samples were defined as c-ILC when all tumor cells were small and uniform with round nuclei and inconspicuous nucleoli (Figure 1B). p-ILC was defined based on previously reported cytological characteristics (i.e., a greater degree of cellular pleomorphism, nuclear membrane irregularities, more prominent nucleoli, increased hyperchromasia, and more frequent mitotic activity) (Figure 1C) [17].
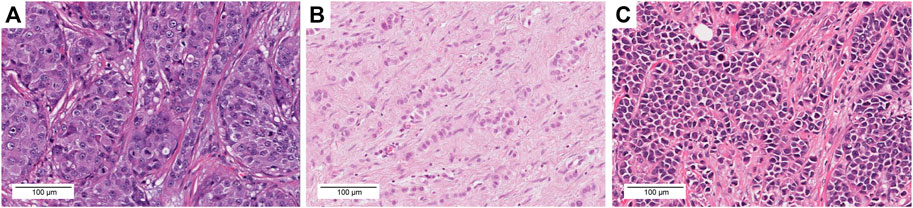
FIGURE 1. Representative images of each histological type with histological grade, molecular subtype, ERBB2 mutation, CDH1 mutation, TMB, and TILs. (A) TCGA-A2-A3XV-01A. H&E images, IBC-NST, GII, Her2 type. The tumor exhibited a solid growth pattern and was composed of tumor cells with severe nuclear atypia. S310F ERBB2 mutation, TMB = 1.8 mut/Mb, TILs = 10%. (B) TCGA-D8-A27G-01A. H&E images, c-ILC, GII, LumA type. This tumor exhibited a linear growth pattern and loose cohesion; tumor cell proliferation was observed along with mild nuclear atypia. I767M ERBB2 mutation, G169Rfs*5 CDH1 mutation, TMB = 31.3 mut/Mb, TILs = 0%. (C) TCGA-BH-A18P-01A. H&E images, p-ILC, GIII, Her2 type. This tumor exhibited a solid growth pattern and loose cohesion; tumor cell proliferation was observed along with severe nuclear atypia. L755S ERBB2 mutation, S36Afs*20 CDH1 mutation, TMB = 11.8 mut/Mb, TILs = 10%. Scale bar = 100 μm, ×20 magnification.
Assessment of TILs
Stromal TILs were quantified on each virtual slide. TILs were assessed in accordance with the guidelines proposed by the international TILs working group [18]. Stromal TILs counts were estimated as the percentage of immune cells in stromal tissues within the tumor. TILs were categorized into three: low (0–10%), intermediate (11–59%), and high (60–100%).
Timer2.0 Database Analysis
To compare ERBB2-mutated BC and ERBB2 non-mutated BC for immune infiltration levels of immune cells including CD8+ T cell, CD4+ T cell, and Treg, the online public resource, Tumor Immune Estimation Resource 2.0 (TIMER2.0; https://timer.cistrome.org/), was utilized [19].
TMB Estimation
TMB is a measure of the total number of mutations per megabase of tumor tissue. It can also be interpreted as the mutation density in tumor genes, defined as the average number of mutations in the tumor genome, including the total number of coding sequence errors, base substitutions, insertion, or deletions [20]. TMB was estimated for TCGA-BRCA (n = 951) and ERBB2-mutated BC (n = 35) and was calculated as the total number of mutations per sample/38, with an estimated exome size of 38 Mb [21]. The samples were classified as TMB-high if they had ≥10 mutations per megabase (mut/Mb) as previously described [22].
Statistical Analysis
The subtype, CDH1 mutation, and TMB between ERBB2 mutated BC and ERBB2 non-mutated BC were analyzed by Fisher exact tests, with a significance threshold of p < 0.05. The correlations for each TMB and TILs were evaluated using the Spearman rank correlation coefficient. Results with p < 0.05 were considered statistically significant.
Results
Clinicopathological Properties for BC With ERBB2 Mutations
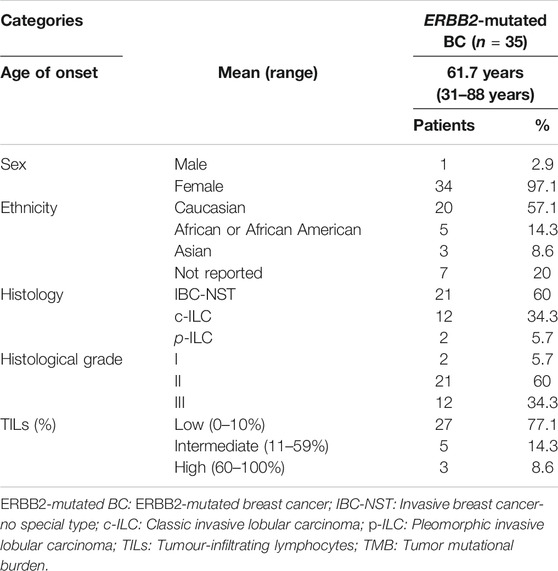
We identified 35 samples of ERBB2-mutated BC among 986 BC samples in TCGA-BRCA (35/986, 3.5%). Two samples (TCGA-A2-A0T6-01A and TCGA-C8-A3M7-01A) exhibited three distinct ERBB2 mutations. Therefore, a total of 39 mutations were identified in 35 samples. The histological types, subtypes, variant types, amino acid changes, mutation sites, TMB, TILs, and CDH1 mutations for each sample are summarized in Supplementary Table S3. The clinicopathological characteristics and TILs for ERBB2-mutated BC are summarized in Table 1. The comparison of ERBB2-mutated BC and ERBB2 non-mutated BC, with respect to subtype, co-occurrence of CDH1 mutation, and TMB, is summarized in Table 2. Briefly, 34 women and 1 man were affected with ERBB2-mutated BC (mean age, 61.7 years; range, 31–88 years). The 35 breast samples corresponded to 21 IBC-NST (60%) cases, 12 c-ILC (34.3%) cases, and two p-ILC (5.7%) cases (Table 2). ERBB2 somatic mutations were detected in 35 samples, and the co-occurrence of ERBB2 and CDH1 mutations was observed in 14 samples (Supplementary Tables S4, S5). CDH1 mutations were observed at significantly higher frequencies (40%; 14/35) in patients with ERBB2 mutated BC than in those with ERBB2 non-mutated BC (Table 2). ERBB2-mutated BCs showed a high histological grade (including a predominantly solid growth pattern, rarely tubule formation, remarkable nuclear atypia, and high mitotic counts) (Figure 1A). The subtypes LumA, LumB, HER2, basal, and normal were detected in 56.3% (18/32), 12.5% (4/32), 18.8% (6/32), 3.1% (1/32), and 9.4% (3/32) of patients, respectively. ERBB2 mutations were present in all subtypes, especially in LumA. However, the subtype proportion between ERBB2 mutated BC and ERBB2 non-mutated BC was not significant. ERBB2-mutated BC had a significantly worse prognosis than ERBB2 non-mutated BC (p < 0.01) (Figure 2).
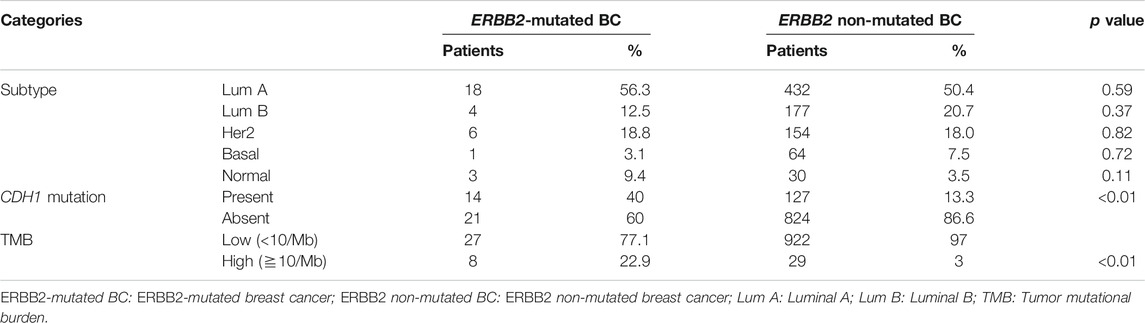
TABLE 2. Comparison of ERBB2-mutated BC and ERBB2 non-mutated BC for subtype and co-occurrence of the CDH1 mutation and TMB.
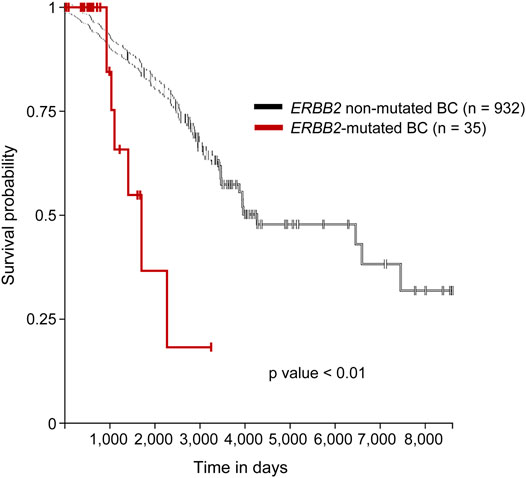
FIGURE 2. Survival analysis of the mutated and non-mutated ERBB2 BC cases. Kaplan–Meier survival curve demonstrated that ERBB2-mutated BC had a significantly worse prognosis than did ERBB2 non-mutated BC. Purple line; ERBB2 mutated BC, black line; ERBB2 non-mutated BC.
Mutation Type and Distribution of ERBB2 Mutation Sites
Of the 39 mutations, 29 were missense mutations (74.4%), four were synonymous mutations (10.2%), two were in-frame insertions (5.1%), one was an in-frame deletion (2.5%), two were present in intronic regions (5.1%) (chr17:g.39712114C > A, chr17:g.39712166C > G), and one was present in the 3′-UTR (2.5%) (chr17:g.39729470G > A).
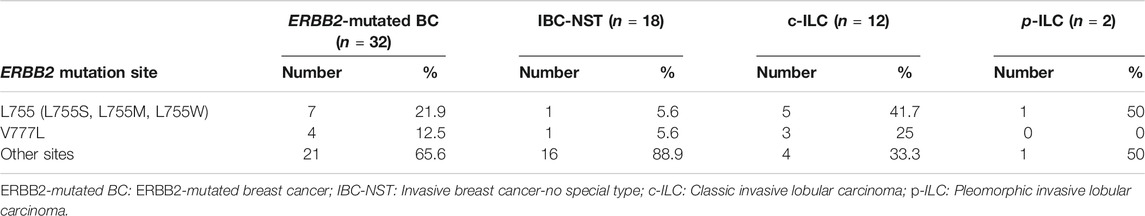
The ERBB2 mutations were associated with 32 types of amino acid changes in the following protein domains/regions: kinase domain (20/32, 62.5%), receptor-L domain (3/32, 9.4%), furin-like cysteine-rich domain (5/32, 15.6%), C-terminal region (2/32, 6.25%), the interdomain region (1/32, 3.1%), and the transmembrane region (1/32, 3.1%) (Figure 3). Substitutions at L755 (L755S, L755M, and L755W) and V777 (V777L) accounted for 34.4% of all amino acid mutations (11/32), especially in patients with c-ILC (66.7%) and p-ILC (50%) (Table 3). In contrast, in IBC-NST, the aforementioned L755 and V777L mutations accounted for approximately 11.2% (2/18) of all mutations (Table 3).
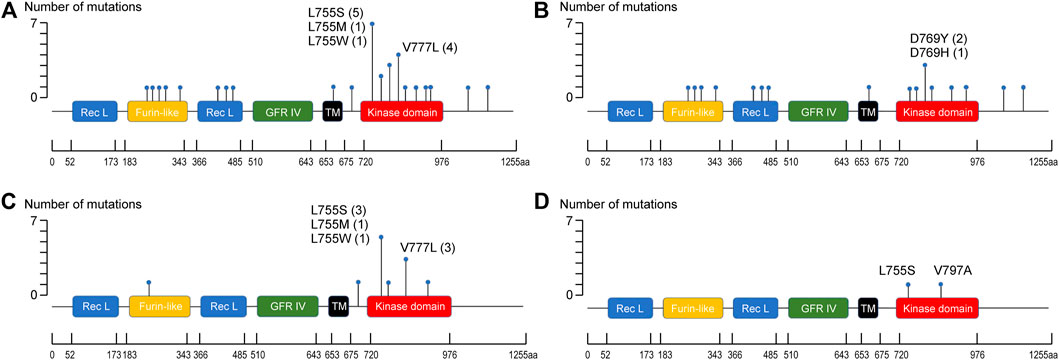
FIGURE 3. Distribution of ERBB2 mutation sites. (A) Distribution of ERBB2 mutation sites in ERBB2-mutated BC. (B) Distribution of ERBB2 mutation sites in ERBB2-mutated IBC-NST. (C) Distribution of ERBB2 mutation sites in ERBB2-mutated c-ILC. (D) Distribution of ERBB2 mutation sites in ERBB2-mutated p-ILC.
Comparison Between the Level of Immune Cell Infiltration in ERBB2-Mutated and Non-mutated BC
CD8+ T cell infiltration (Figure 4A) was significantly higher in ERBB2-mutated BC than in their non-mutant counterparts (p < 0.05). In contrast, no differences in CD4+ T cell and Treg infiltration were observed between ERBB2 mutated and non-mutated BC (Figures 4B,C), as determined by TIMER and CIBERSORT analyses using TIMER2.0.
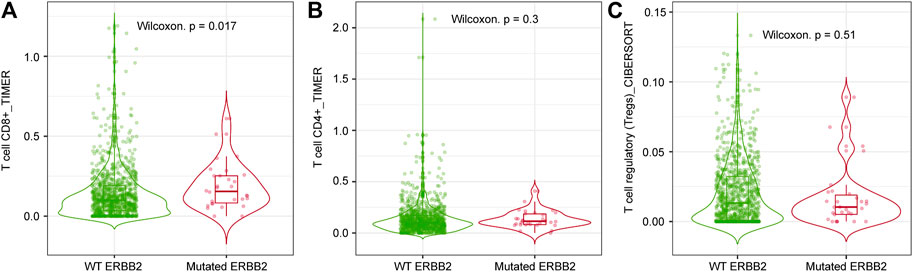
FIGURE 4. Comparison between the level of immune infiltration levels in ERBB2-mutated and non-mutated BC via TIMER2.0 analysis. (A) CD8+ T cell infiltration level in ERBB2-mutated and non-mutated BC (TIMER analysis). (B) CD4+ T cell infiltration level in ERBB2-mutated and non-mutated BC (TIMER analysis). (C) Treg infiltration level in ERBB2-mutated and non-mutated BC (CIBERSORT analysis).
TMB and TILs in ERBB2-Mutated BC
The mean TMB in ERBB2-mutant BC was 13.6 mut/Mb (median = 3.0 mut/Mb), while the mean TMB in the TCGA-BRCA cohort without ERBB2 mutations (n = 951) was 3.2 mut/Mb (median = 1.8 mut/Mb). The TMB in ERBB2-mutated BC was significantly higher than that ERBB2 non-mutated BC (p < 0.01) (Table 2; Figure 5). Furthermore, 3.0% (29/951) of the samples in the ERBB2 non-mutated BC were classified as TMB-high based on the calculated cut-off of 10 mut/Mb. In comparison, 22.9% (8/35) of the ERBB2-mutated BC samples were TMB-high. Furthermore, 8.6% (3/35) of ERBB2-mutated BC samples were TILs-high, 14.3% (5/35) were TIL-intermediate, and 77.1% (27/35) were TILs-low, including 12 samples with no TILs (34.3%). The correlation coefficient for the relationship between TMB and TILs was 0.38 (Spearman's rank correlation, p < 0.05), indicating a weak positive correlation (Figures 6, 7).
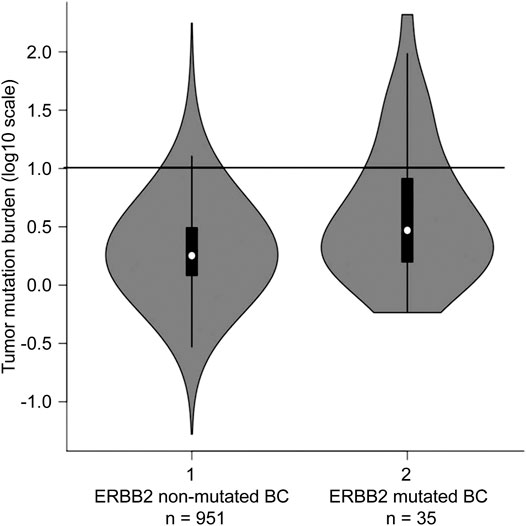
FIGURE 5. Tumour mutational burden (TMB) in ERBB2 non-mutated BC (n = 951) and ERBB2 mutated BC (n = 35) The black line indicating 10 mutations/megabase represents the threshold for TMB-high. For ERBB2 non-mutated BC, the frequency of TMB-high was 3.0% (29/951); for ERBB2-mutated BC, the frequency was 22.9% (8/35).
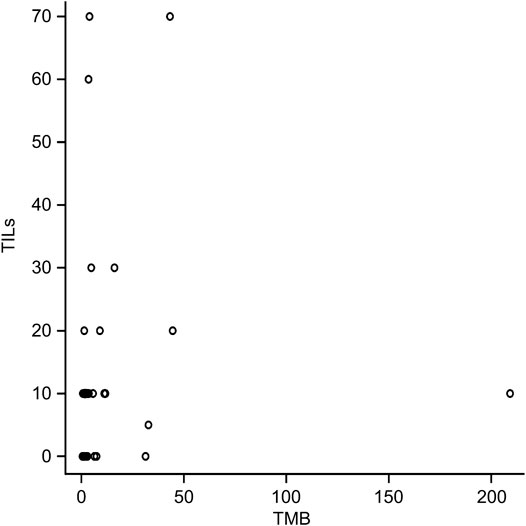
FIGURE 6. Correlation between tumor mutational burden (TMB) and tumour-infiltrating lymphocytes (TILs). Spearman’s rank correlation coefficient, 0.38 (p < 0.05).
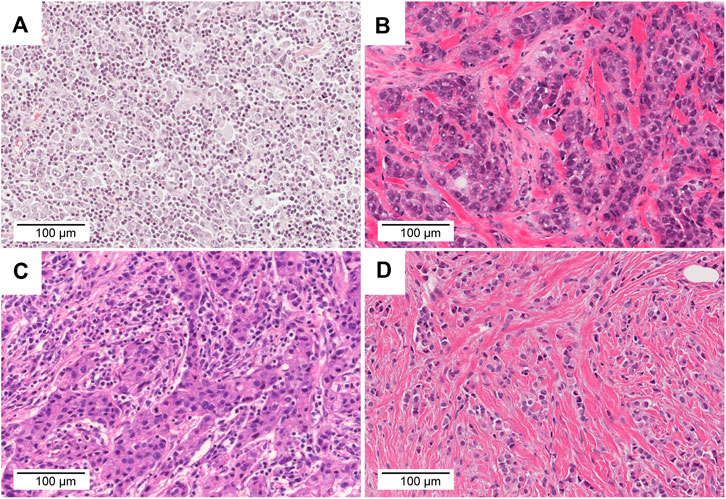
FIGURE 7. Comparison of TMB, TILs, and histology. (A) TCGA-AO-A128-01A. This tumour was classified as TMB-high and TIL-high (TMB = 43.3 mut/Mb, TILs = 70%). (B) TCGA-AN-A46-01A. This tumor was classified as TMB-high and TIL-low (TMB = 209.2 mut/Mb, TILs = 10%). (C) TCGA-OL-A66P-01A. This tumor was classified as TMB-low and TIL-high (TMB = 4.0 mut/Mb, TILs = 70%). (D) TCGA-AC-A3YI-01A. This tumor was classified as TMB-low and TIL-low (TMB = 0.6 mut/Mb, TILs = 0%). Scale bar = 100 μm, ×20 magnification.
Discussion
We characterized the clinicopathological features, immune infiltration levels, TMB (mut/Mb), and TILs (%) associated with ERBB2-mutated BC using bioinformatics. ERBB2-mutated BC was identified in 3.5% of TCGA-BRCA samples, concurrent with a previous report [3]. In this study, among the 35 samples, 60% were IBC-NST and 40% were ILC (c-ILC, p-ILC). As ILC typically accounts for 5–15% of all BCs [15], ILC appears to be enriched in ERBB2 mutations. A similar trend has been reported previously [4, 7]. In ERBB2-mutated BC, ILC is frequently related to a high frequency of CDH1 mutations. In this study, 40% (14/35) of the ERBB2 mutant cases harbored CDH1 mutation.
In our study, missense mutations in the kinase domain were the most common ERBB2 mutation type in BC. The mutation type and site differ among tumor types. For example, ERBB2 mutations in micropapillary urothelial carcinoma of the urinary bladder are predominantly present in the extracellular domain [23].
In this study, L755 mutations and V777L accounted for 34.4% of all ERBB2 mutations in BC. Remarkably, more than half of c-ILC and p-ILC samples revealed the presence of amino acid change at L755 and V777L. A correlation has been reported between p-ILC and amino acid change at L755 [14]; however, in this study, a similar correlation was also observed for c-ILC. Together with previous reports [14, 24], our study shows that kinase domain (predominantly L755 site and V777L substation, especially in lobular carcinoma) is a hot spot for ERBB2 mutation in BC. A previous study integrated three cohorts (METABRIC, TCGA, MSK-IMPACT) and calculated the frequency of the ERBB2 mutation (2.2%, 34/1,580). The ERBB2 mutation was enriched in ILC with a prevalence of 5.7% (n = 16) vs. 1.4% in IDC (n = 18) [25]. Based on these findings, it is thought that an analysis of other data sets (METABRIC and MSK-IMPACT) will yield the same results.
Previous studies have reported that irreversible tyrosine kinase inhibitors (TKIs, such as neratinib and afatinib) are useful for treating ERBB2-mutated BC [1, 26, 27]. Ongoing clinical trials are investigating the effect of neratinib (SUMMIT trial; NCT01953926) and afatinib (NCI-MATCH; NCT02465060) in patients with ERBB2-mutant cancers. However, neratinib-resistant BC has been previously reported [26]. Acquired tolerance to TKIs occurs via multiple mechanisms, including gatekeeper mutations and “bypass” resistance [28]. The acquisition of therapeutic resistance in cancer cells is a major challenge of molecular targeted therapy. Therefore, we evaluated TMB and TILs to investigate the possibility of immunotherapy as an alternative treatment, including the use of TKIs, such as neratinib and afatinib.
TMB is a predictive biomarker for the response to immune checkpoint inhibitor (ICI) therapy, and some clinical studies have reported a response to immunotherapy based on a high TMB [29, 30]. In fact, in June 2020, the US-FDA approved pembrolizumab for treating solid tumors in adults and children with unresectable or metastatic high TMB (≥10 mut/Mb) solid tumors based on the results of the KEYNOTE-158 trial [31]. In the present study, we found that the TMB is significantly higher in ERBB2-mutated BC than in ERBB2 non-mutated BC samples without ERBB2 mutations (p < 0.01). In BC, the TMB is usually lower (2.6 mut/Mb) than in other carcinomas, such as lung cancer [21, 22]. In a previous study based on 3,969 primary and metastatic BC samples, approximately 5% of the samples were classified as TMB-high [22]. However, in this study, 22.9% of the ERBB2-mutated BC samples were TMB-high, and ERBB2-mutated BC was regarded as a subset of TMB-high BC. TILs are a favorable prognostic factor and are associated with the response to PD-1/PD-L1 inhibitors in BC [32]. The samples used in our study exhibited the following TILs frequencies: high (8.6%; 3/35); intermediate (14.3%; 5/35), and low (77.1%; 27/35), including 12 samples with no TILs (34.3%). In a previous study, 44.2% of the samples were TILs-low, 36.3% were TILs-intermediate, and 19.2% were TIL-high [33]. In another study, TILs were lacking in 16% of the samples [34]. In our study, ERBB2-mutated BC was characterized by low TILs count. TILs are reportedly higher in triple-negative and Her2-positive BC subtypes than in the luminal subtype. Moreover, ILCs are TILs-low compared with other histological types [33]. The TILs pattern in ERBB2-mutated BC resembled the pattern observed in hormone receptor-positive/Her2-negative BC and ILC [34]. Although ERBB2-mutated BCs are classified as TMB-high and TILs-low, a positive correlation was observed between TMB and TILs in this study. TIMER2.0 analysis revealed that the CD8+ T infiltration level of ERBB2-mutated BC was significantly higher than that of ERBB2 non-mutated BC. In ERBB2-mutated BC, the CD8 + T cell infiltration level was considered to be up-regulated, and subpopulations of TILs may contain much CD8+ T cells. However, further investigations are needed to confirm this.
This study demonstrated that 22.9% of the ERBB2-mutated BCs were TMB-high BC; in contrast, 77.1% of them were TILs-low. From this result, it is questionable whether ERBB2-mutated cases would really benefit from ICI therapy. Further research is needed to determine whether it is an indication for ICI therapy. Since TMB estimation are highly variable [35], with slower turn-around times and high costs, further studies are required to validate the correlations with between other biomarkers, such as PD-L1 immunostaining and microsatellite instability (MSI), and TMB. A recent study reported that a machine learning algorithm, Image2TMB, can predict the TMB directly from images of H&E-stained histopathological sections [36]; however, this method is still not used in the clinical setting, although it may be a novel method of estimating the TMB easily.
This study has some limitations. This result was obtained from one dataset (TCGA-BRCA), and it is necessary to verify the obtained result by laboratory research in the future.
In conclusion, The TMB and CD8+ T cell infiltration level in ERBB2-mutated BC samples was significantly higher than that in ERBB2 non-mutated BC. Additionally, 22.9% of the ERBB2-mutated BC samples were identified as TMB-high, and a positive correlation was identified between TMB and TILs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SH and TS designed the experiments and performed the pathological reassessment. TK performed the statistical analyses, SU and TK analyzed the data. All authors were involved in writing the paper and approved of the submitted manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Bioinformatics Consultation Forum, The Society for Biotechnology, Japan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2021.633243/full#supplementary-material.
References
1. Bose, R, Kavuri, SM, Searleman, AC, Shen, W, Shen, D, Koboldt, DC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov (2013) 3:224–37. doi:10.1158/2159-8290.cd-12-0349
2. Zabransky, DJ, Yankaskas, CL, Cochran, RL, Wong, HY, Croessmann, S, Chu, D, et al. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci USA (2015) 112:E6205–E6214. doi:10.1073/pnas.1516853112
3. Petrelli, F, Tomasello, G, Barni, S, Lonati, V, Passalacqua, R, and Ghidini, M. Clinical and pathological characterization of HER2 mutations in human breast cancer: a systematic review of the literature. Breast Cancer Res Treat (2017) 166:339–49. doi:10.1007/s10549-017-4419-x
4. Ma, CX, Bose, R, Gao, F, Freedman, RA, Telli, ML, Kimmick, G, et al. Neratinib efficacy and circulating tumor DNA detection of HER2 mutations in HER2 nonamplified metastatic breast cancer. Clin Cancer Res (2017) 23:5687–95. doi:10.1158/1078-0432.ccr-17-0900
5. Berx, G, Cleton-Jansen, AM, Nollet, F, De Leeuw, WJ, Van de Vijver, M, Cornelisse, C, et al. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J (1995) 14:6107–15. doi:10.1002/j.1460-2075.1995.tb00301.x
6. Takeichi, M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science (1991) 251:1451–5. doi:10.1126/science.2006419
7. Ross, JS, Wang, K, Sheehan, CE, Boguniewicz, AB, Otto, G, Downing, SR, et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin Cancer Res (2013) 19:2668–76. doi:10.1158/1078-0432.ccr-13-0295
8. Wang, T, Xu, Y, Sheng, S, Yuan, H, Ouyang, T, Li, J, et al. HER2somatic mutations are associated with poor survival in HER2-negative breast cancers. Cancer Sci (2017) 108:671–7. doi:10.1111/cas.13182
9. Ping, Z, Siegal, GP, Harada, S, Eltoum, I-E, Youssef, M, Shen, T, et al. ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget (2016) 7:80655–63. doi:10.18632/oncotarget.13019
10. Christgen, M, Bartels, S, Luft, A, Persing, S, Henkel, D, Lehmann, U, et al. Activating human epidermal growth factor receptor 2 (HER2) gene mutation in bone metastases from breast cancer. Virchows Arch (2018) 473:577–82. doi:10.1007/s00428-018-2414-1
11. Lien, H-C, Chen, Y-L, Juang, Y-L, and Jeng, Y-M Frequent alterations of HER2 through mutation, amplification, or overexpression in pleomorphic lobular carcinoma of the breast. Breast Cancer Res Treat (2015) 150:447–55. doi:10.1007/s10549-015-3336-0
12. Christgen, M, Bartels, S, Radner, M, Raap, M, Rieger, L, Christgen, H, et al. ERBB2 mutation frequency in lobular breast cancer with pleomorphic histology or high-risk characteristics by molecular expression profiling. Genes Chromosomes Cancer (2019) 58:175–85. doi:10.1002/gcc.22716
13. Deniziaut, G, Tille, JC, Bidard, F-C, Vacher, S, Schnitzler, A, Chemlali, W, et al. ERBB2 mutations associated with solid variant of high-grade invasive lobular breast carcinomas. Oncotarget (2016) 7:73337–46. doi:10.18632/oncotarget.11819
14. Ding, Q, Chen, H, Lim, B, Damodaran, S, Chen, W, Tripathy, D, et al. HER2 somatic mutation analysis in breast cancer: correlation with clinicopathological features. Hum Pathol (2019) 92:32–8. doi:10.1016/j.humpath.2019.07.006
15.WHO Classification of Tumours Editorial Board. WHO classification of tumours. In: Breast tumours. 5th ed. Lyon: IARC (2019).
16. Elston, CW, and Ellis, IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology (1991) 19:403–10. doi:10.1111/j.1365-2559.1991.tb00229.x
17. Weidner, N, and Semple, JP. Pleomorphic variant of invasive lobular carcinoma of the breast. Hum Pathol (1992) 23:1167–71. doi:10.1016/0046-8177(92)90035-2
18. Salgado, R, Denkert, C, Demaria, S, Sirtaine, N, Klauschen, F, Pruneri, G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol (2015) 26:259–71. doi:10.1093/annonc/mdu450
19. Li, T, Fu, J, Zeng, Z, Cohen, D, Li, J, Chen, Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucl. Acids Res (2020) 48:W509–W514. doi:10.1093/nar/gkaa407
20. Schumacher, TN, Kesmir, C, and van Buuren, MM Biomarkers in cancer immunotherapy. Cancer Cell (2015) 27:12–4. doi:10.1016/j.ccell.2014.12.004
21. Chalmers, ZR, Connelly, CF, Fabrizio, D, Gay, L, Ali, SM, Ennis, R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med (2017) 9:34. doi:10.1186/s13073-017-0424-2
22. Barroso-Sousa, R, Jain, E, Cohen, O, Kim, D, Buendia-Buendia, J, Winer, E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol (2020) 31:387–94. doi:10.1016/j.annonc.2019.11.010
23. Ross, JS, Wang, K, Gay, LM, Al-Rohil, RN, Nazeer, T, Sheehan, CE, et al. A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin Cancer Res (2014) 20:68–75. doi:10.1158/1078-0432.ccr-13-1992
24. Ross, JS, Gay, LM, Wang, K, Ali, SM, Chumsri, S, Elvin, JA, et al. NonamplificationERBB2genomic alterations in 5605 cases of recurrent and metastatic breast cancer: an emerging opportunity for anti-HER2 targeted therapies. Cancer (2016) 122:2654–62. doi:10.1002/cncr.30102
25. Kurozumi, S, Alsaleem, M, Monteiro, CJ, Bhardwaj, K, Joosten, SEP, Fujii, T, et al. Targetable ERBB2 mutation status is an independent marker of adverse prognosis in estrogen receptor positive, ERBB2 non-amplified primary lobular breast carcinoma: a retrospective in silico analysis of public datasets. Breast Cancer Res (2020) 22:85. doi:10.1186/s13058-020-01324-4
26. Ben-Baruch, NE, Bose, R, Kavuri, SM, Ma, CX, and Ellis, MJ. HER2-Mutated breast cancer responds to treatment with single-agent neratinib, a second-generation HER2/EGFR tyrosine kinase inhibitor. J Natl Compr Canc Netw (2015) 13:1061–4. doi:10.6004/jnccn.2015.0131
27. Hanker, AB, Brewer, MR, Sheehan, JH, Koch, JP, Sliwoski, GR, Nagy, R, et al. An acquired HER2T798I gatekeeper mutation induces resistance to neratinib in a patient with HER2 mutant-driven breast cancer. Cancer Discov (2017) 7:575–85. doi:10.1158/2159-8290.cd-16-1431
28. Westover, D, Zugazagoitia, J, Cho, BC, Lovly, CM, and Paz-Ares, L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol (2018) 29:i10–i19. doi:10.1093/annonc/mdx703
29. Wu, Y, Xu, J, Du, C, Wu, Y, Xia, D, Lv, W, et al. The predictive value of tumor mutation burden on efficacy of immune checkpoint inhibitors in cancers: a systematic review and meta-analysis. Front Oncol (2019) 9:1161. doi:10.3389/fonc.2019.01161
30. Samstein, RM, Lee, C-H, Shoushtari, AN, Hellmann, MD, Shen, R, Janjigian, YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet (2019) 51:202–6. doi:10.1038/s41588-018-0312-8
31.FDA. FDA approves pembrolizumab for adults and children with TMB-H solid tumors (2020). http://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors. doi:10.31525/fda1-ucm626720.htm
32. Gonzalez-Ericsson, PI, Stovgaard, ES, Sua, LF, Reisenbichler, E, Kos, Z, Carter, JM, et al. The path to a better biomarker: application of a risk management framework for the implementation of PD-L1 and TILs as immuno-oncology biomarkers in breast cancer clinical trials and daily practice. J Pathol (2020) 250:667–84. doi:10.1002/path.5406
33. Denkert, C, von Minckwitz, G, Darb-Esfahani, S, Lederer, B, Heppner, BI, Weber, KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol (2018) 19:40–50. doi:10.1016/s1470-2045(17)30904-x
34. Stanton, SE, Adams, S, and Disis, ML. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes. JAMA Oncol (2016) 2:1354–60. doi:10.1001/jamaoncol.2016.1061
35. Merino, DM, McShane, L, Butler, M, Funari, VA, Hellmann, MD, Chaudhary, R, et al. TMB standardization by alignment to reference standards: phase II of the friends of cancer research TMB harmonization project. J Clin Oncol (2019) 37:2631. doi:10.1200/jco.2019.37.15_suppl.2624
Keywords: Breast cancer, ErbB2 mutation, tumor mutational burden, tumour-infiltrating lymphocytes, Bioinformactics analysis
Citation: Uchida S, Kojima T and Sugino T (2021) Clinicopathological Features, Tumor Mutational Burden, and Tumour-Infiltrating Lymphocyte Interplay in ERBB2-Mutated Breast Cancer: In Silico Analysis. Pathol. Oncol. Res. 27:633243. doi: 10.3389/pore.2021.633243
Received: 25 November 2020; Accepted: 01 March 2021;
Published: 01 April 2021.
Edited by:
Anna Sebestyén, Semmelweis University, HungaryCopyright © 2021 Uchida, Kojima and Sugino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiro Uchida, RHIuVWNoaWRhQGdtYWlsLmNvbQ==
 Shiro Uchida1,2,3*
Shiro Uchida1,2,3*