- 1Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Dali University, Dali, China
- 2Institute of Translational Medicine for Metabolic Diseases, Dali University, Dali, China
The epidemic of the novel, pathogenic SARS-coronavirus 2 (SARS-CoV-2) in the world pose a global health emergency. Cancer has been identified as a risk factor for the novel Coronavirus disease 2019 (COVID-19). The ACE2 and TMPRSS2 have been implicated in SARS-CoV-2 infection for mediating viral entry into the host cell. However, a systematic analysis of aberrant expression of ACE2 and TMPRSS2 was not yet reported in multiple human cancers. Here, we analyzed gene expression of ACE2 and TMPRSS2 across 31 types of tumors. Notably, overexpression of ACE2 and TMPRSS2 have been observed in colorectal cancer including colon adenocarcinoma (COAD), and rectum adenocarcinoma (READ). In addition, the colorectal tumors with upregulated gene expressing presented with decreased DNA methylation levels. DNA methylation might be one of the reasons for abnormal expression of ACE2 and TMPRSS2. Conclusively, colorectal cancer was the only cancer with the upregulated expression of ACE2 and TMPRSS2. More care of colorectal cancer patients is needed in multiple cancers affected by the COVID-19 outbreak.
Introduction
The novel Coronavirus disease 2019 (COVID-19) is the global health emergency [1]. To date (October 1, 2020), COVID-19 has resulted in more than 37 million human infections. As the number of infected people increases, early identification of high-risk individuals is an urgent medical need. The angiotensin I converting enzyme 2 (ACE2) has been implicated in SARS-CoV-2 infection for mediating viral entry into the host cell [2]. ACE2 is the main receptor for the spike (S) protein of SARS-CoV-2, mediating viral attachment to target cells. The transmembrane serine protease 2 (TMPRSS2) has also been proposed to modulate susceptibility to SARS-CoV-2 [3]. TMPRSS2 is a serine protease that cleaves SARS-CoV-2 spike protein, facilitating viral entry and activation. If a cell expresses both ACE2 and TMPRSS2, it has a higher risk of infection by SARS-CoV-2 [4].
Cancer has been already identified as an individual risk factor for COVID-19 [5]. We conducted a pan-cancer analysis of ACE2 and TMPRSS2 across 31 types of tumors in TCGA pan-cancer datasets. The susceptibility and symptoms of different cancer patients were systematically assessed by analyzing abnormal expression of ACE2 and TMPRSS2.
Method
GEPIA2 (http://gepia2.cancer-pku.cn) is an online dataset for analyzing RNA sequencing expression data from the TCGA projects. It provides tumor/normal differential expression analysis according to cancer types. Expression of ACE2 and TMPRSS2 were compared in tumor from TCGA pan-cancer datasets and its normal control tissue from GTEx datasets using GEPIA2 database [6]. In this study, |log2FC)| > 1 and p < 0.01 were selected as the standard for different expression.
cBioPortal (http://www.cbioportal.org), a comprehensive web resource, can visualize and analyze multidimensional cancer genomics data. The DNA copy number variation of ACE2 and TMPRSS2 had been analyzed by cBioportal database tool [7]. UALCAN (http://ualcan.path.uab.edu) provides methylation analyses based on TCGA cohort data. In our study, the promoter methylation levels of these two genes in different cancers were examined using UALCAN database [8]. The p value cutoff was 0.05.
Results
We compared expression of ACE2 and TMPRSS2 in tumor and its normal control tissue in TCGA pan-cancer datasets using GEPIA2 (Supplementary Figures, S1, S2). ACE2 expression was upregulated in five tumors. Notably, colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRP), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ) and stomach adenocarcinoma (STAD) presented with increased ACE2 expression (Figure 1A). TMPRSS2 expression was upregulated in seven tumors, including cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, kidney chromophobe (KICH), prostate adenocarcinoma (PRAD), READ, uterine corpus endometrial carcinoma (UCEC) and uterine carcinosarcoma (UCS) (Figure 1B). From the results above, it can be seen that two cancers, COAD and READ, highly express both ACE2 and TMPRSS2.
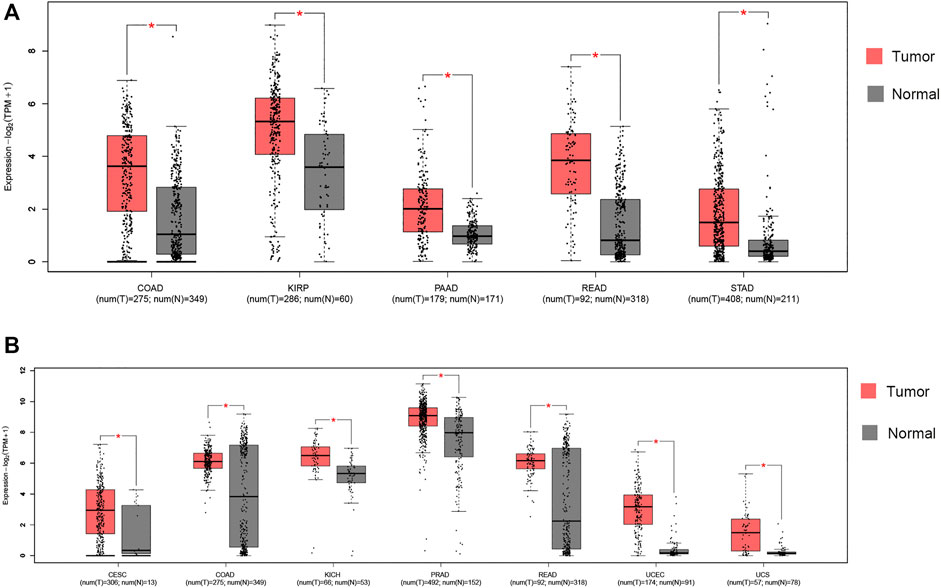
FIGURE 1. RNA expression of ACE2 and TMPRSS2 in tumors. (A) Colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRP), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ) and stomach adenocarcinoma (STAD) presented increased ACE2 expression. *|log2FC)| > 1 and p < 0.01. These data were obtained using GEPIA2. Red, tumor samples; gray, normal samples. (B) Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, kidney chromophobe (KICH), prostate adenocarcinoma (PRAD), READ, uterine corpus endometrial carcinoma (UCEC) and uterine carcinosarcoma (UCS) presented increased TMPRSS2 expression. *|log2FC)| > 1 and p < 0.01. These data were obtained using GEPIA2. Red, tumor samples; gray, normal samples.
To investigate the cause of the upregulated expression in colorectal cancer, we analyzed the DNA copy number variation of ACE2 and TMPRSS2. There were less than 1% patients with ACE2 amplification, and no patient had TMPRSS2 amplification in colorectal cancer (Figure 2A). Thus, it is possible that the upregulation of ACE2 and TMPRSS2 expression was resulted from epigenetic regulation. We then examined the promoter methylation levels of these two genes in colorectal cancer using UALCAN database. We have found that the tumors with upregulated gene expressing presented with decreased DNA methylation levels of ACE2 and TMPRSS2, including COAD and READ (Figures 2B,C). Our findings suggested that DNA methylation might be one of the reasons of abnormal expression of ACE2 and TMPRSS2.
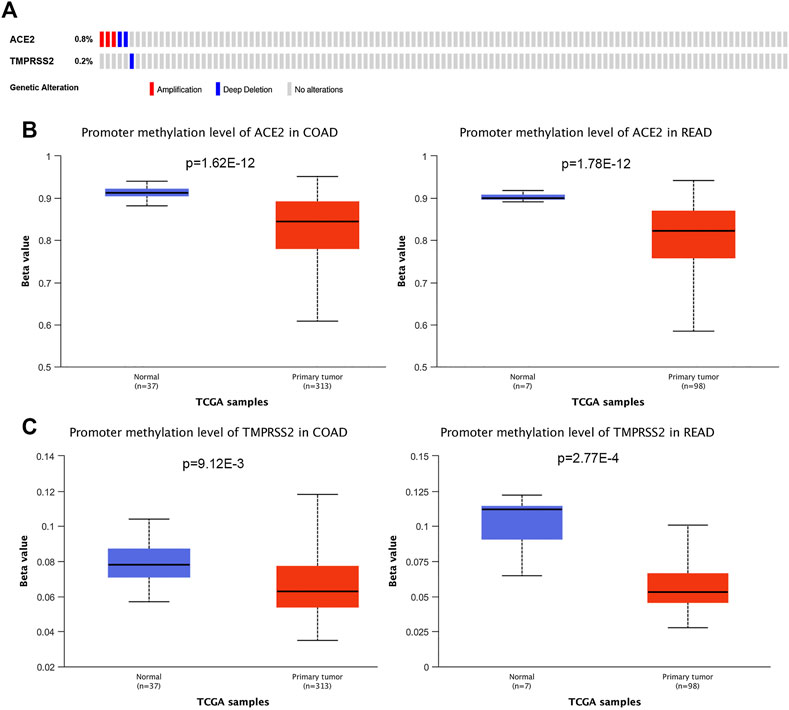
FIGURE 2. Genetic alteration and DNA methylation of ACE2 and TMPRSS2 in tumors. (A) DNA copy number variations of ACE2 and TMPRSS2 across the colorectal cancer samples (COAD and READ). The mutation mapper tool of cBioPortal was used to plot the distribution of mutations. (B) DNA methylation aberration of ACE2 in COAD and READ. This data was obtained using Ualcan. (C) DNA methylation aberration of TMPRSS2 in COAD and READ. This data was obtained using Ualcan.
Discussion
Patients with cancer have been disproportionally affected by the COVID-19 pandemic. Cancer has been identified as an individual risk factor for COVID-19 [5]. In patients with cancer, COVID-19 can be especially harsh. This is likely because many patients have a weakened immune system [9]. Certain types of cancer itself can increase a risk of getting an infection. From our pan-cancer analysis, it can be seen that two cancers, COAD and READ, highly express both ACE2 and TMPRSS2. Both cancers occur in the colorectum of the digestive tract. Notably, researchers have found that the coronavirus SARS-CoV-2 can infect cells of the intestine, which could explain the observation that approximately one third of COVID-19 patients experience gastrointestinal symptoms such as diarrhea, and the fact that the virus often can be detected in stool samples [10]. Therefore, patients with colorectal cancer may have more severe gastrointestinal symptoms.
To investigate the cause of the upregulated expression in colorectal cancer, we analyzed the DNA copy number variation and promoter methylation level of ACE2 and TMPRSS2. We found DNA copy number variations were not relevant to expression of ACE2 and TMPRSS2. We then examined the promoter methylation levels of these two genes in colorectal cancer. We have found that the tumors with upregulated gene expressing presented with decreased DNA methylation levels of ACE2 and TMPRSS2, including COAD and READ. Our findings suggested that DNA methylation might be the reason of abnormal expression of ACE2 and TMPRSS2. Other mechanisms of transcriptional regulation may also have an impact on the abnormal expression of ACE2 and TMPRSS2, which requires further explorations.
In summary, among many types of cancers, colorectal cancer was the only one with the upregulated expression of ACE2 and TMPRSS2. Moreover, it has been proved that the SARS-CoV-2 can infect cells of the intestine. Therefore, patients with colorectal cancer may have more severe gastrointestinal symptoms. Our results suggest a need for more care of colorectal cancer patients in areas affected by the COVID-19 outbreak. Since our study is a preliminary analysis, further validation is warranted.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
HW and JY designed and performed the experiments and drafted the manuscript; HW was responsible for the sample collection and data analysis; HW and JY discussed, revised and approved the manuscript. All the authors approved this manuscript.
Funding
This study was supported by grants from National Natural Science Foundation of China (81760565), and Youth Top Talent project of High-level talent development support program of Yunnan Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2021.612969/full#supplementary-material.
References
1. Phan, T. Novel coronavirus: from discovery to clinical diagnostics. Infect Genet Evol (2020). 79:104211. doi:10.1016/j.meegid.2020.104211
2. Zhou, P, Yang, X-L, Wang, X-G, Hu, B, Zhang, L, Zhang, W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020). 579:270–3. doi:10.1038/s41586-020-2012-7
3. Hoffmann, M, Kleine-Weber, H, Schroeder, S, Krüger, N, Herrler, T, Erichsen, S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell (2020). 181:271–80. doi:10.1016/j.cell.2020.02.052
4. Ziegler, CGK, Allon, SJ, Nyquist, SK, Mbano, IM, Miao, VN, Tzouanas, CN, et al. HCA Lung Biological Network. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020). 181:1016–35. doi:10.1016/j.cell.2020.04.035
5. Liang, W, Guan, W, Chen, R, Wang, W, Li, J, Xu, K, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol (2020). 21:335–7. doi:10.1016/s1470-2045(20)30096-6
6. Tang, Z, Kang, B, Li, C, Chen, T, and Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res (2019). 47:W556–W560. doi:10.1093/nar/gkz430
7. Gao, J, Aksoy, BA, Dogrusoz, U, Dresdner, G, Gross, B, Sumer, SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling (2013). 6:pl1. doi:10.1126/scisignal.2004088
8. Chandrashekar, DS, Bashel, B, Balasubramanya, SAH, Creighton, CJ, Ponce-Rodriguez, I, Chakravarthi, BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017). 19:649–58. doi:10.1016/j.neo.2017.05.002
9. Bakouny, Z, Hawley, JE, Choueiri, TK, Peters, S, Rini, BI, Warner, JL, et al. COVID-19 and cancer: current challenges and perspectives. Cancer Cel (2020). 38(5):629–46. 10.1016/j.ccell.2020.09
Keywords: SARS-COV-2, cancer, ACE2, TMPRSS2, colorectum
Citation: Wang H and Yang J (2021) Colorectal Cancer that Highly Express Both ACE2 and TMPRSS2, Suggesting Severe Symptoms to SARS-CoV-2 Infection. Pathol. Oncol. Res. 27:612969. doi: 10.3389/pore.2021.612969
Received: 01 October 2020; Accepted: 26 February 2021;
Published: 15 April 2021.
Edited by:
József Tímár, Semmelweis University, HungaryCopyright © 2021 Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiankang Yang, amt5YW5nMTk4NEAxMjYuY29t
 Huai Wang1,2
Huai Wang1,2