Abstract
Nucleotide-binding and leucine-rich repeat protein (NLRP) genes are involved in inflammasome formation that plays a role in inflammation/host defense and cell death. Both cell death and inflammation are crucial for cancer development, but the roles of NLRPs in cancer are partially known. In this study, we analyzed mononucleotide repeats in coding sequences of NLRP1, NLRP2, NLRP4 and NLRP9, and found 1, 1, 1 and 8 frameshift mutation (s) in gastric (GC) and colonic cancers (CRC), respectively. Five of the 32 high microsatellite instability (MSI-H) GCs (15.5%) and 6 of 113 MSI-H CRCs (5.5%) exhibited the frameshift mutations. There was no NLRP frameshift mutations in microsatellite stable (MSS) GCs and CRCs. We also discovered that 2 of 16 CRCs (12.5%) harbored intratumoral heterogeneity (ITH) of the NLRP9 frameshift mutations in one or more areas. In both GC and CRC with MSI-H, NLRP9 expression in NLRP9-mutated cases was significantly lower than that in NLRP9-non-mutated cases. Our data indicate that NLRP9 is altered at multiple levels (frameshift mutation, mutational ITH and loss of expression), which together could contribute to pathogenesis of MSI-H GC and CRC.
Introduction
Nucleotide-binding and leucine-rich repeat proteins (NLRPs), also known as NALPs, are crucial mediators in inflammation and host defense [1–3]. Currently, there are 10 known human NLRP genes (NLRP1-6, 9, 10, 12, 14). They, in common, possess pyrin, NACHT and leucine-rich repeat domains and are crucial for aggregating other proteins that form the inflammasome [1–3]. Of the 10 NLRP genes, gene functions of NLRP1, 3 and 5 have been known well [4–6]. NLRP3 inflammasome formation enables activation of caspase-1 and subsequent interleukin-1β and interleukin-18 activation, which could develop cell death and inflammation [7]. Both cell death and inflammation are crucial for cancer development, but the roles of NLRPs in cancer development remain controversial due to the diverse cancer-related findings [1, 5]. For example, recent studies identified NLRPs that modulated the mucosal immune response during inflammatory bowel disease-associated tumorigenesis [8, 9]. NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis [9]. In skin squamous cell carcinoma, NLRP1 inflammasome pathway is silenced [10]. Also, NLRP2 inhibits cell proliferation and tumor growth of human glioblastoma [11]. NLRC3, a putative NLR, has anti-cancerous functions [12]. These data suggest tumor suppressor gene (TSG) functions of NLRPs in cancer development. By contrast, tumor-promoting (oncogenic) functions of NLRPs have been discovered in many cancers as well. NLRP1 overexpression is correlated with the tumorigenesis and proliferation of human breast cancer [13]. In melanoma, NLRP1 promotes tumor growth by enhancing inflammasome activation and suppressing apoptosis [14]. Activation of NLRP3 inflammasome promotes inflammation-induced tumor growth and metastasis in many cancers [2]. These data may indicate that NLRPs are involved in cancer pathogenesis, but their cancer-related alterations vary depending on cancer types.
DNA mismatch repair (MMR) is a cellular mechanism for correcting erroneous bases by MMR-specific proteins, alterations of which would result in microsatellite instability (MSI) and mutator phenotypes [15]. The mutator phenotype is characterized by mutation aCRCumulation in repetitive DNA sequences (frequently mono- or dinucleotide repeats). In coding DNA sequences, the MSI produces frameshift mutations within the affected proteins that would truncate protein synthesis [16]. Gastric (GC), colonic (CRC) and endometrial cancers are the most common cancers with high MSI (MSI-H) phenotype [17]. It is believed that MSI is random, but there is evidence suggesting that MSI targets include a growing list of cancer genes such as TGF-β1 gene and BAX gene [18, 19]. There are nucleotide repeats in coding DNA sequences of NLRP1, 2, 4 and 9, which could be altered in MSI cancers. In the present study, we detected frequent NLRP1, 2, 4 and 9 frameshift mutations in GC and CRC with MSI-H.
Materials and Methods
Cancer Tissues
In the present study, formalin-fixed and paraffin embedded (FFPE) tissues of 235 cancers from Korean patients (77 GCs and 158 CRCs) were used (Table 1). Briefly, they consisted of 32 GCs with MSI-H, 45 GCs with microsatellite stable (MSS), 113 CRCs with MSI-H and 45 CRCs with MSS. For the evaluation of the MSI status of each cancer, we adopted five mononucleotide repeats (BAT25, BAT26, NR-21, NR-24 and MONO-27) that were known to be frequently mutated in MSI-H cancers [20]. Malignant cells and normal cells were separately collected from hematoxylin-eosin slides using a 30G1/2 hypodermic needle by microdissection as described in earlier studies [21, 22]. DNA extraction was performed by a modified single-step DNA extraction method using proteinase K [21, 22]. Research approval was obtained from the institutional review board of Catholic University of Korea. All FFPE samples were made anonymous and waived the need for written informed consent.
TABLE 1
| Feature | MSI-H | MSS |
|---|---|---|
| Gastric carcinomas | ||
| Total cases | 32 | 45 |
| TNM stage | ||
| I | 11 | 15 |
| II | 13 | 18 |
| III | 7 | 11 |
| IV | 1 | 1 |
| Colon carcinomas | ||
| Total cases | 113 | 45 |
| TNM stage | ||
| I | 20 | 6 |
| II | 50 | 20 |
| III | 37 | 16 |
| IV | 6 | 3 |
| Location | ||
| Cecum | 33 | 0 |
| Ascending colon | 58 | 3 |
| Transverse colon | 15 | 2 |
| Descending & sigmoid colon | 6 | 17 |
| Rectum | 1 | 23 |
Summary of pathologic features of gastric and colon cancers.
TNM: tumor, lymph node, metastasis, MSI-H: high microsatellite instability, MSS: stable microsatellite instability.
TNM stage is defined by AJCC 8th edition.
Mutational Analysis
There is one A7 (exon 4; primers 5′-AAGCTCAGCCATTGGGACC-3′, 5′- AAGGTGGAGATGATGGCCC-3′) in NLRP1 gene, one A7 (exon 13; primers 5′-TTTCTTCCCCCATTGTACCCC-3′, 5′- TCTGCCCAGGGATGATGTTTC-3′) in NLRP2 gene, one T7 (exon 2; primers 5′- TCACCCAGCTGTGAGATGTG-3′, 5′- TCTTGGGACAGTTGGAAGCC-3′) in NLRP4 gene, and one A7 (exon 2; primers 5′-TGAGCGATGATTGGAGGCAG-3′, 5′-GAGTTTTGGATGCCGCAACA-3′) and one T8 (exon 1; primers 5′- CTTTTC CCTCTGGAGACACCTC-3′, 5′-TTCTCCAAAGGTTGTTTGAGGA-3′) in NLRP9 gene. Genomic DNA from the microdissected cells was amplified by polymerase chain reaction (PCR) using the primer pairs. Radioisotope ([32P]dCTP) was incorporated into the PCR products for detection by autoradiogram. For the screening of the mutations, aberrant gel motility in single strand conformation polymorphism (SSCP) was used (FMC Mutation Detection Enhancement system; Intermountain Scientific, Kaysville, UT, United States) [21, 22]. Cancer DNA with mobility shifts in the SSCP was subsequently sequenced by Sanger DNA sequencing of both forward and reverse strands to confirm the mutated sequences (3730 DNA Analyzer, Applied Biosystem, Carlsbad, CA, United States). We also analyzed intratumoral heterogeneity (ITH) of the NLRP9 mutations, which could be altered in MSI-H cases. For this, 16 MSI-H CRCs with 4–7 different areas per CRC were studied by PCR-SSCP and Sanger sequencing as described above.
Expressional Analysis
Since frameshift mutations of genes in MSI-H frequently accompany expressional alteration of the affected proteins [21], we analyzed the NLRP9 protein expression status in the GCs and CRCs by immunohistochemistry using anti-NLRP9 antibody (catalogue number HPA042623, Atlas Antibodies, Stockholm, Sweden; dilution 1/50). The immunohistochemistry procedures have been described in our earlier studies [21]. Briefly, sections from FFPE GC and CRC tissues were studied using ImmPRESS System (Vector Laboratories, Burlingame, CA, United States). After deparaffinization, heat-induced epitope retrieval was conducted by immersing the slides in Coplin jars filled with 10 mmol/L citrate buffer (pH 6.0) and boiling the buffer for 30 min in a pressure cooker (Nordic Ware, Minneapolis, MN) inside a microwave oven at 700 W; the jars were then cooled for 20 min. We used diaminobenzidine (brown) as chromogen for the immunohistochemistry reactions and counterstained with hematoxylin (blue). The staining intensity was graded as follows: 0, negative; 1+, weak staining in cytosol or nucleus; 2+, moderate; and 3+, intense. The extent was graded as follows: 0, 0–5% positivity of cells; 1, 6–19%; 2, 20–49%; 3, >50%. The intensity and the extent were multiplied for the immunohistochemistry score (IS), which consisted of IS 0–2 as −, 3 or 4 as + and 6 or 9 as ++. Negative control of the immunostaining was the replacement of primary antibody with the blocking reagent.
Results
NLRP Gene Mutations
The PCR-SSCP for the mononucleotide repeats in NLRP1, 2, 4 and 9 showed aberrant migrating bands in 5 GCs and 6 CRCs, which were subsequently confirmed as frameshift mutations (deletion or duplication mutation of one base within the nucleotide repeats) by DNA sequencing (Table 2 and Figure 1). Eight, one, one and one mutation (s) were discovered in NLRP9, NLRP1, NLRP2, and NLRP4, respectively (Table 2). Of note, all of the cancers with the NLRP frameshift mutations were MSI-H cases, but was none in the MSS cases (significant difference, p = 0.004). These cancers harbored one NLRP frameshift mutation in each cancer without harboring any other mutation among these genes. All the mutations were interpreted somatic as there was no such mutations in matched normal tissues. The DNA sequencing exhibited both wild-type and mutation sequences, indicating they were heterozygous mutations (Figures 1, 2). Five of the 32 MSI-H GCs (15.5%) and 6 of 113 MSI-H CRCs (5.5%) exhibited the frameshift mutations with no significant organ difference (p > 0.05). There was no significant difference of 5-years survival between patients with and without the mutations (p > 0.05).
TABLE 2
| Gene | Wild type | Mutation | MSI status of the mutation cases (n) | Incidence in MSI-H cancers (%) | Nucleotide change (predicted amino acid change) |
|---|---|---|---|---|---|
| NLRP9 | T8 | T9 | MSI-H (1) | Gastric: 1/32 (3.1) | c.19dupT (p.Ser7PhefsX21) |
| T7 | MSI-H (5) | Gastric: 2/32 (6.3) | c.19delT (p.Ser7ArgfsX9) | ||
| Colon: 3/113 (2.7) | |||||
| A7 | A6 | MSI-H (2) | Gastric: 1/32 (3.1) | c.791delA (p.Lys264ArgfsX12) | |
| Colon: 1/113 (0.9) | |||||
| NLRP1 | A7 | A6 | MSI-H (1) | Colon: 1/113 (0.9) | c.1748delA (p.Lys583ArgfsX14) |
| NLRP2 | A7 | A6 | MSI-H (1) | Colon: 1/113 (0.9) | c.3113delA (p.Asn1038ThrfsX4) |
| NLRP4 | T7 | T6 | MSI-H (1) | Gastric: 1/32 (3.1) | c.1496delT (p.Leu499TrpfsX5) |
NLRP mutations in gastric and colon cancers.
MSI-H: high microsatellite instability.
FIGURE 1
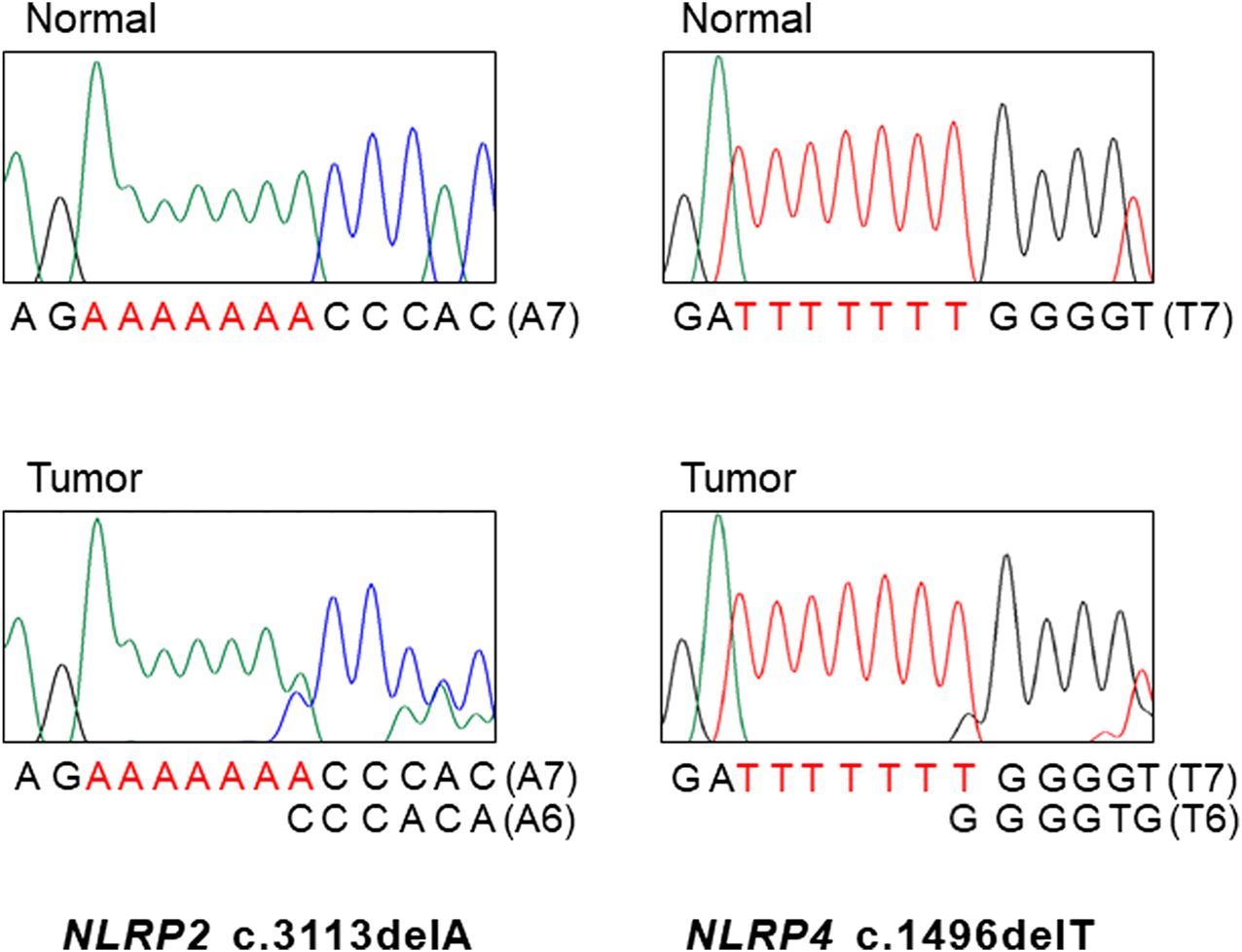
DNA sequencings NLRP2 and NLRP4 mutations in gastric and colon cancers. DNA sequencing analyses of the A7 repeat (left) of NLRP2 and the T7 repeat (right) of NLRP4 from normal (upper) and tumor tissues (lower). Sanger DNA sequencing analyses reveal heterozygous deletions of a base within the repeats in the tumor tissues as compared to normal tissues.
FIGURE 2
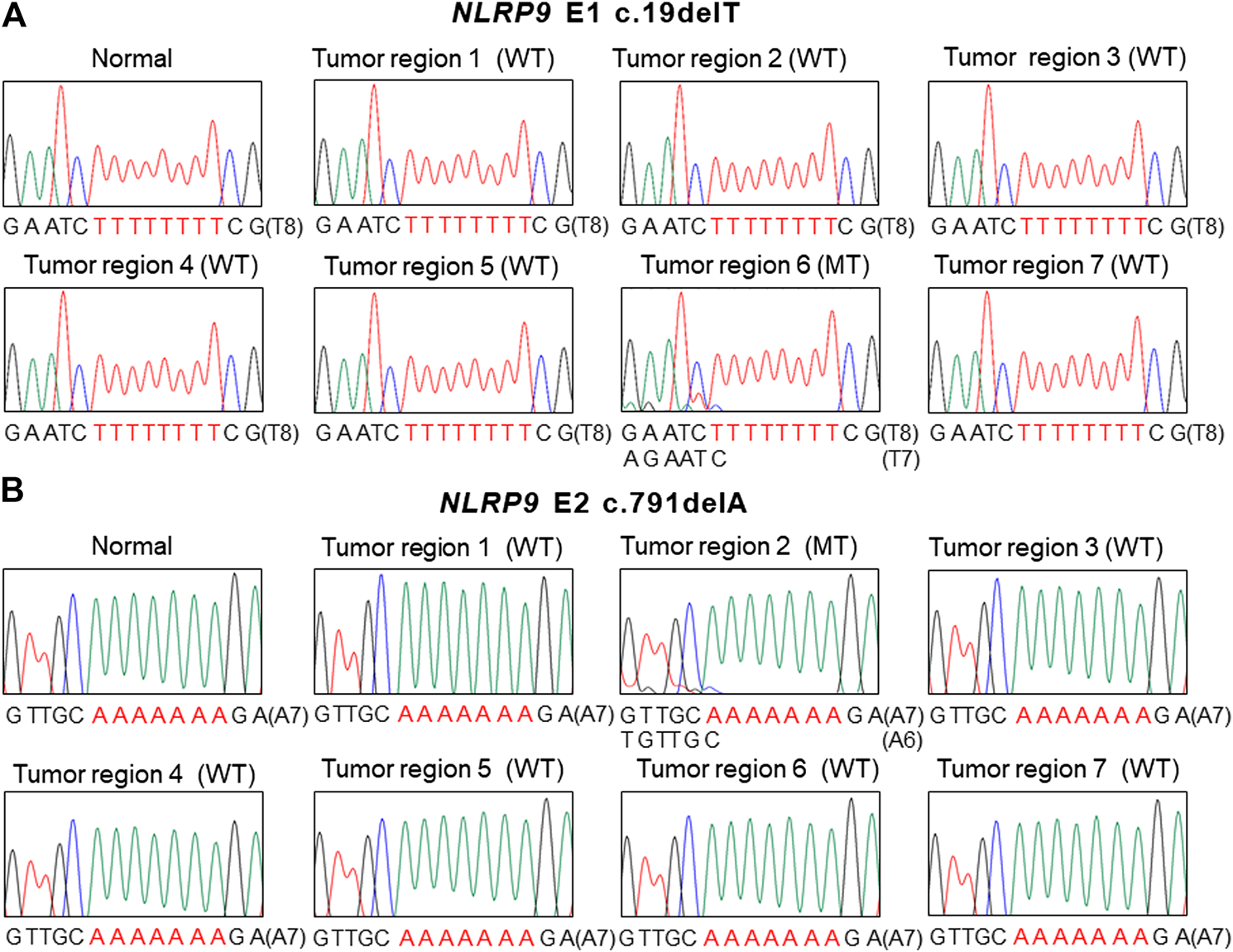
Intratumoral heterogeneity of NLRP9 frameshift mutation in colon cancer. (A) Sanger DNA sequencing shows NLRP9 c.19delT mutation (MT) in one region (6) and wild-type (WT) in the other 6 regions (1, 2, 3, 4, 5, 7). (B. Sanger DNA sequencing shows NLRP9 c.791delA mutation (MT) in one region (2) and wild-type (WT) in the other 6 regions (1, 3, 4, 5, 6, 7).
For the multiregional mutation analysis (4–7 areas), 2 of 16 CRCs analyzed (12.5%) revealed the NLRP9 frameshift mutations in one region and the wild-type in the other 6 regions (Figure 2). However, we were not able to find histologic differences between the ITH lesions examined by a pathologist. Also, there was no significant difference in patients’ survival nor clinical outcomes between ITH (n = 2) and non-ITH (n = 14) cases (p > 0.05). We analyzed the ITH of NLRP1, NLRP2 and NLRP4 frameshift mutation, but did not find any ITH of them.
Immunohistochemistry
We next studied the expression status of NLRP9 protein since the NLRP9 mutation was most common among the NLRP genes analyzed in this study. In normal gastric and colonic mucosa, NLRP9 was well expressed (IS 6 or 9) by immunohistochemistry (Figures 3A,D). The positive immunostaining was mainly detected in cytosol (Figure 3). The negative control using blocking reagent instead of the primary antibody showed no immunostainings in the tissues. In cancers, the MSS (71.1%, 64/90) and MSI-H (64.8%, 94/145) cancers exhibited no statistically different prevalence in NLRP9 expression (Fisher’s exact test, p = 0.197) (Table 3). In both GC and CRC with MSI-H, NLRP9 expression in NLRP9-mutated cases was significantly lower than that in NLRP9-non-mutated cases (p = 0.009 and p = 0.016, respectively). There was no significant difference in NLRP9 expression between GC and CRC with MSI-H (p > 0.05).
FIGURE 3
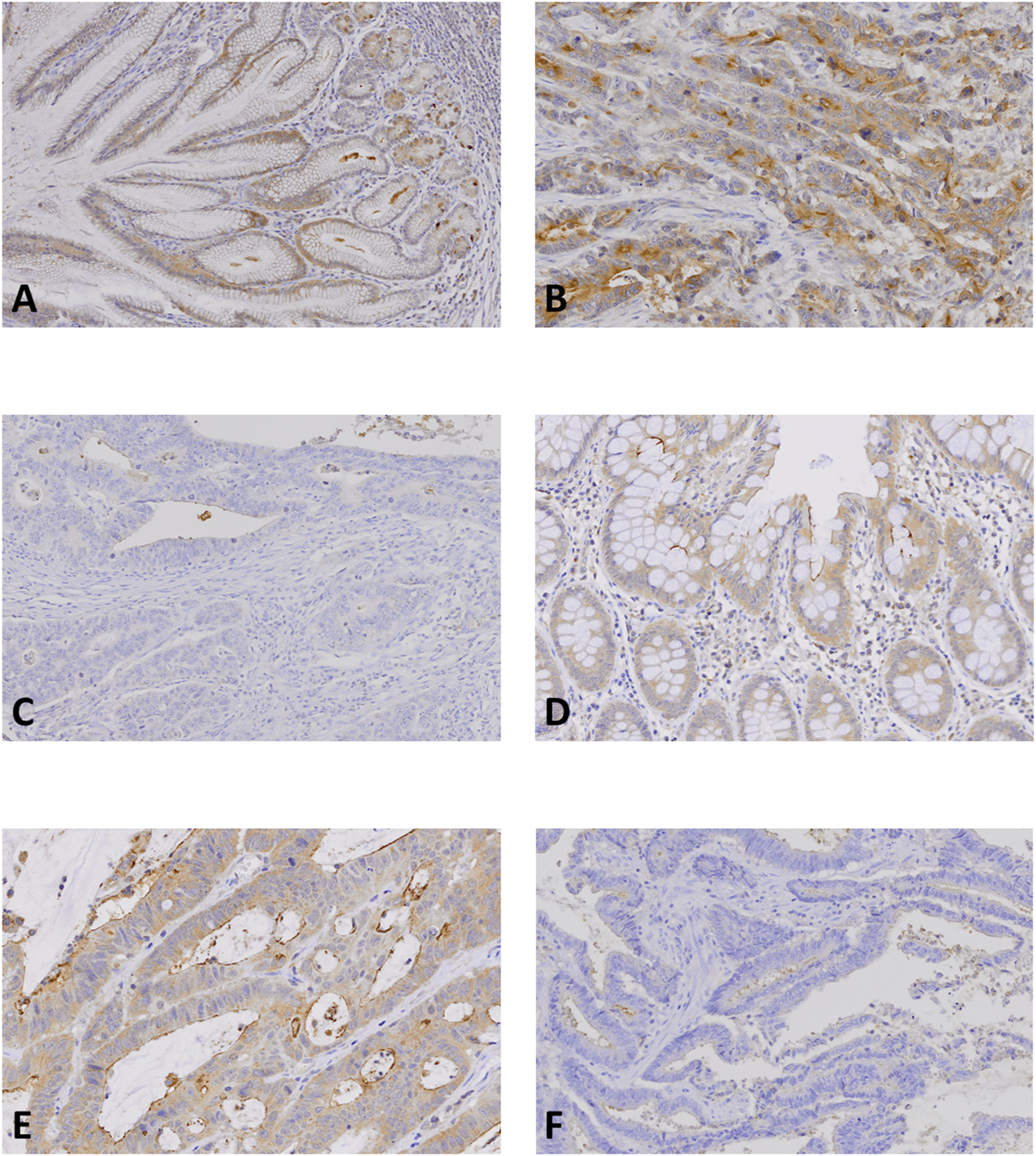
NLRP9 expression in gastric and colon cancer tissues. (A, D): Normal gastric (A) and colonic (D) mucosal epithelial cells show positive NLRP9 immunostaining. (B, E): Gastric (B) and colon (E) cancers show positive NLRP9 immunostaining in the cancer cells. (C, F): In gastric (C) and colon cancers (F) without the NLRP9 frameshift mutations, the cancer cells show negative NLRP9 immunostaining.
TABLE 3
| All cases | Positive expression (+, ++) | p-value | |
|---|---|---|---|
| MSI-H | |||
| Gastric cancer | |||
| Total | 32 | 21 (+: 7, ++:14) | 0.009* |
| With NLRP9 mutation | 4 | 0 (+:0, ++:0) | |
| Without NLRP9 mutation | 28 | 21 (+:7, ++:14) | |
| Colon cancer | |||
| Total | 113 | 73 (+: 23, ++:50) | 0.016* |
| With NLRP9 mutation | 4 | 0 (+:0, ++:0) | |
| Without NLRP9 mutation | 109 | 72 (+:22, ++:50) | |
| MSS | |||
| Gastric cancer | |||
| Total | 45 | 32 (+:10, ++:22) | |
| without NLRP9 mutation | 45 | 32 (+:10, ++:22) | |
| Colon cancer | |||
| Total | 45 | 30 (+:7, ++:23) | |
| without NLRP9 mutation | 45 | 30 (+:7, ++:23) |
NLRP9 expression in gastric and colon cancers.
GC: gastric cancer, CRC: coloic cancer, MSI-H: high microsatellite instability, MSS: stable microsatellite instability; * significant.
Discussion
The role of inflammation is well-known in both development and progression of cancers, and is a hallmark in cancer development [23]. Also, evasion of cell death is known to play an important role in the pathogenesis of cancers [23]. In this sense, the inflammasome associated with both inflammation and cell death could be a good candidate for the cancer research. To find cancer-related alterations in cancer inflammasome, we analyzed frameshift mutations of NLRP (NLRP1, 2, 4 and 9) genes closely related to the inflammasome in GC and CRC in this study. The mutations were frequent in both MSI-H GC (15.5%) and CRC (5.5%); NLRP9 mutations revealed ITH; and NLRP9 expression was frequently lost in NLRP9-mutated cancers. These data indicate that NLRPs, especially NLRP9, might be involved in MSI-H cancer pathogenesis. In general, a cancer-related gene may exhibit not only a high incidence of genetic alterations but also functional consequences [23]. For NLRP9 gene, until now its role has been suggested only in maturation and fertilization of gonocytes [24]. Neither cancer-related functions nor gastrointestinal functions of NLRP9 gene have been identified. Mouse orthologue Nlrp9b is specifically expressed in intestinal epithelial cells and restricts rotavirus infection by activating pyroptosis [25]. It can be speculated that the NLRP9 frameshift mutations could inactivate the pyroptosis. However, relationship between pyroptosis and MSI cancers is not known currently. As The Human Protein Atlas database (https://www.proteinatlas.org/) exhibits that NLRP9 expression is evident in normal stomach and colon cells and is strongest in colorectal cancers followed by gastric, pancreatic, testicular and liver cancers, but other cancer tissues are in general weakly stained or negative, suggesting its role in gastrointestinal tract. Together, these data suggest that NLRP9 expression is common in both normal and cancer cells of stomach and colon, and that frameshift mutations and expression loss of NLRP9 is frequent in GC and CRC with MSI-H. However, it remains to be clarified whether these alterations have a causative role in tumorigenesis, or they simply reflect the phenomenon that MSI-H cancers have an increased frequency of mutations.
In addition to NLRP9, we found NLRP1, 2 and 4 mutations in GC and CRC with MSI-H with lower incidences (Table 2). As mentioned above [9, 13], NLRP1 possesses both oncogenic and TSG activities. Also, NLRP2 revealed TSG functions in glioblastoma and oncogenic functions in bladder cancer [26]. These opposing phenomenon may preclude predicting functional consequences of the frameshift mutations. NLRP4 negatively regulates autophagy processes through an association with beclin1 [27]. There is controversy about the roles of autophagy in cancer (tumor suppressing vs. promoting) [28]. The low incidence as well as the debate suggests that the NLRP4 mutation may not necessary play an important role in cancer development.
NLRP9 expression was negative in all cancers with its frameshift mutations (Table 3). Anti-NLRP9 antibody adopted in this study was made using a synthetic peptide of amino acids 472-549 (isoform 1). Based on the nucleotide changes of NLRP9 by the frameshift mutations, 2 amino acid locations (amino acids 7 and 264) would be altered and truncated (Table 2). Parts of pyrin domain (amino acids 1-94), NACHT domain (146-465) and leucine repeat domains (743-907) (www.uniprot.org) would be truncated by the mutations and thus the truncated area may not be detected by the immunohistochemistry.
Our data indicate that the mutational ITH of NLRP9 is common in the CRC (12.5% of MSI-H CRCs) (Figure 2). ITH is important in driving phenotypic selection and adaptation in response to selective pressures in a cancer [29]. ITH is known to be associated with poor prognosis and clinical outcomes [29]. The ITH in our study could ameliorate the phenotypic consequences of the mutation and further functional and clinical implication of the ITH is required to be defined.
Statements
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by the institutional review board of Catholic University of Korea.
Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
SL and NY designed the study: SM and HS performed the experiment; HM and MW analyzed the data.
Funding
This study was supported by grants from Korea Research Foundation (2019R1A5A2027588 and 2020R1A2C2005031).
Acknowledgments
This study was supported by grants from Korea Research Foundation (2019R1A5A2027588 and 2020R1A2C2005031).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Martinon F Tschopp J . Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ (2007). 14:10–22. 10.1038/sj.cdd.4402038
2.
Karan D . Inflammasomes: emerging central players in cancer immunology and immunotherapy. Front Immunol (2018). 9:3028. 10.3389/fimmu.2018.03028
3.
Place DE Kanneganti T-D . Recent advances in inflammasome biology. Curr Opin Immunol (2018). 50:32–8. 10.1016/j.coi.2017.10.011
4.
Chavarría-Smith J Vance RE . The NLRP1 inflammasomes. Immunol Rev (2015). 265:22–34. 10.1111/imr.12283
5.
Moossavi M Parsamanesh N Bahrami A Atkin SL Sahebkar A . Role of the NLRP3 inflammasome in cancer. Mol Cancer (2018). 17:158. 10.1186/s12943-018-0900-3
6.
Alimohammadi M Björklund P Hallgren Å Pöntynen N Szinnai G Shikama N et al Autoimmune polyendocrine syndrome type 1 and NALP5, a parathyroid autoantigen. N Engl J Med (2008). 358:1018–28. 10.1056/nejmoa0706487
7.
Franchi L Eigenbrod T Muñoz-Planillo R Nuñez G . The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol (2009). 10:241–7. 10.1038/ni.1703
8.
Allen IC TeKippe EM Woodford R-MT Uronis JM Holl EK Rogers AB et al The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med (2010). 207:1045–56. 10.1084/jem.20100050
9.
Williams TM Leeth RA Rothschild DE Coutermarsh-Ott SL McDaniel DK Simmons AE et al The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol (2015). 194:3369–80. 10.4049/jimmunol.1402098
10.
Sand J Fenini G Grossi S Hennig P Di Filippo M Levesque M et al The NLRP1 inflammasome pathway is silenced in cutaneous squamous cell carcinoma. J Invest Dermatol (2019). 139:1788–97. 10.1016/j.jid.2019.01.025
11.
Wu G-Q Liao Y-J Qin Z-Q He L-R Chen Y-C Zeng Y-X et al PYRIN domain of NALP2 inhibits cell proliferation and tumor growth of human glioblastoma. Plasmid (2010). 64:41–50. 10.1016/j.plasmid.2010.01.003
12.
Karki R Man SM Malireddi RKS Kesavardhana S Zhu Q Burton AR et al NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature (2016). 540:583–7. 10.1038/nature20597
13.
Wei Y Huang H Qiu Z Li H Tan J Ren G et al NLRP1 overexpression is correlated with the tumorigenesis and proliferation of human breast tumor. Biomed Res Int (2017). 2017:4938473. 10.1155/2017/4938473
14.
Zhai Z Liu W Kaur M Luo Y Domenico J Samson JM et al NLRP1 promotes tumor growth by enhancing inflammasome activation and suppressing apoptosis in metastatic melanoma. Oncogene (2017). 36:3820–30. 10.1038/onc.2017.26
15.
Iyer RR Pluciennik A Burdett V Modrich PL . DNA mismatch repair: functions and mechanisms. Chem Rev (2006). 106:302–23. 10.1021/cr0404794
16.
Imai K Yamamoto H . Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis (2008). 29:673–80. 10.1093/carcin/bgm228
17.
Kim T-M Laird PW Park PJ . The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell (2013). 155:858–68. 10.1016/j.cell.2013.10.015
18.
Ionov Y Yamamoto H Krajewski S Reed JC Perucho M . Mutational inactivation of the proapoptotic gene bax confers selective advantage during tumor clonal evolution. Proc Natl Acad Sci (2000). 97:10872–7. 10.1073/pnas.190210897
19.
Markowitz S Wang J Myeroff L Parsons R Sun L Lutterbaugh J et al Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science (1995). 268:1336–8. 10.1126/science.7761852
20.
Murphy KM Zhang S Geiger T Hafez MJ Bacher J Berg KD et al Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn (2006). 8:305–11. 10.2353/jmoldx.2006.050092
21.
Mo HY An CH Choi EJ Yoo NJ Lee SH . Somatic mutation and loss of expression of a candidate tumor suppressor gene TET3 in gastric and colorectal cancers. Pathol Res Pract (2020). 216:152759. 10.1016/j.prp.2019.152759
22.
An CH Je EM Yoo NJ Lee SH . Frameshift mutations of cadherin genes DCHS2, CDH10 and CDH24 genes in gastric and colorectal cancers with high microsatellite instability. Pathol Oncol Res (2015). 21:181–5. 10.1007/s12253-014-9804-8
23.
Hanahan D Weinberg RA . Hallmarks of cancer: the next generation. Cell (2011). 144:646–74. 10.1016/j.cell.2011.02.013
24.
Dalbiès-Tran R Papillier P Pennetier S Uzbekova S Monget P . Bovine mater-like NALP9 is an oocyte marker gene. Mol Reprod Dev (2005). 71:414–21. 10.1002/mrd.20298
25.
Zhu S Ding S Wang P Wei Z Pan W Palm NW et al Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature (2017). 546:667–70. 10.1038/nature22967
26.
Poli G Brancorsini S Cochetti G Barillaro F Egidi MG Mearini E . Mearini, Expression of inflammasome-related genes in bladder cancer and their association with cytokeratin 20 messenger RNA. Urol Oncol (2015). 33:505. 10.1016/j.urolonc.2015.07.012
27.
Jounai N Kobiyama K Shiina M Ogata K Ishii KJ Takeshita F . NLRP4 negatively regulates autophagic processes through an association with beclin1. J.I. (2011). 186:1646–55. 10.4049/jimmunol.1001654
28.
Ahn CH Jeong EG Lee JW Kim MS Kim SH Kim SS et al Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS (2007). 115:1344–9. 10.1111/j.1600-0463.2007.00858.x
29.
Ramón Y Cajal S Sesé M Capdevila C Aasen T De Mattos-Arruda L et al Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berlin) (2020). 98:161–77. 10.1007/s00109-020-01874-2
Summary
Keywords
NLRP, Somatic mutation, loss of expression, cancer, colon cancer
Citation
Moon SW, Son HJ, Mo HY, Yoo NJ and Lee SH (2021) Somatic Mutation of NLRP Genes in Gastric and Colonic Cancers. Pathol. Oncol. Res. 27:607385. doi: 10.3389/pore.2021.607385
Received
17 September 2020
Accepted
26 February 2021
Published
16 April 2021
Volume
27 - 2021
Edited by
László Kopper, Semmelweis University, Hungary
Updates
Copyright
© 2021 Moon, Son, Mo, Yoo and Lee.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sug Hyung Lee, suhulee@catholic.ac.kr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.