Abstract
Objective: Although polymorphisms of adiponectin gene (ADIPOQ) in obesity-related conditions have been the target of research efforts, little is known about this genetic marker in uterine leiomyomas. The aim of this pilot study was to analyze the frequencies of alleles and genotypes of Single Nucleotide Polymorphism ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) and to correlate it with the risk of uterine fibroids.
Study Design: The Test Group comprised 90 women treated surgically for uterine leiomyomas in the Department of Operative Gynecology, Endoscopy and Gynecologic Oncology, Polish Mother’s Memorial Hospital-Research Institute. 90 disease-free individuals were used as Controls. Patients within both groups were additionally stratified into lean, overweight and obese, according to Body Mass Index. Statistical analysis was performed between the two major groups and, furthermore, within the abovementioned subgroups.
Results: The study revealed no statistically significant differences in the distribution of alleles and genotypes of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) between the two main groups. A weak correlation within distributions of alleles was observed between obese Test Patients and lean Controls.
Conclusion: This pilot study has revealed no association between SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) and uterine fibroids. Further studies on larger groups are warranted to elucidate whether this SNP may be correlated with uterine leiomyomas.
Introduction
Uterine fibroids (UFs), often referred to as uterine leiomyomas, are highly prevalent benign smooth muscle tumors of the uterus. Lifetime risk of this condition varies according to sources, but may reach as high as 75% [1, 2]. Numerous theories have been proposed on how and why UFs develop, however, their pathogenesis has not been yet clarified satisfactorily and is not fully understood [1–3]. Several studies have shown that the prevalence of UF is highest in females aged 26–30 and directly corresponds to overall body adiposity expressed by Body Mass Index (BMI) [4, 5]. The latter correlation is consistent with estrogen-dependency doctrine, which has gained particular attention, where excess fatty tissue eventually leads to elevated estrogen levels by facilitating peripheral conversion of androgen to estrogen.
Regrettably, not much scientific attention has been focused on understanding genetics and epigenetics of UFs. Välimäki et al. [6] however, have published a large study on more than 15,000 uterine fibroids cases and almost 400,000 controls. The authors have pointed 22 loci which displayed a genome-wide significant susceptibility to leiomyomagenesis. These genes were linked with two distinct biological processes: genome stability and genitourinary development. In other studies, it has been suggested, that the vast majority of UFs can be subject to one of four major categories of mutations: MED 12 mutations, FH inactivation, COL4A6-COL4A5 deletions, or HMGA2 overexpression [7–9]. However, still little is known about genetic polymorphism in UFs.
On the contrary, widespread research efforts have been put into exploring obesity and genetic polymorphism in obesity-related genes. This trend could be explained by the modern approach to adipose tissue, which is not only the energy storage compartment but rather a vital endocrine organ [10, 11]. Among adipokines, two major players have gained particular attention: adiponectin and leptin with approximately 8,500 and 16,000 titles in PubMed database, respectively (as of October 2021). Adiponectin, which is synthesized only in adipocytes, is inversely correlated with overall body fat content [12, 14] and exerts a protective influence on such conditions as: diabetes, insulin resistance or even endometrial cancer [13–19].
The issue of genetic polymorphism in adiponectin gene and its association with obesity has been widely studied. Synonymous Single Nucleotide Polymorphism ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) has been thoroughly analyzed in up-to-date literature on the abovementioned matter and its correlation with BMI is generally hypothesized [20–25]. The findings are however incoherent probably due to various factors including ethnicity and geographical distribution of tested subjects [26–28]. Moreover, the frequency of genetic variants also differs across populations: Minor Allele Frequency (MAF) reaches from 0, 15 to 0, 34 [29]. Although the exact biological significance of this SNP and the specific mechanisms how its variants alter the levels and activity of adiponectin remain unclear, it is postulated that this marker may potentially affect transcriptional activity or splicing efficiency [30].
In our earlier study [32] in which we analyzed the role of ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in Endometrial Cancer, patients treated for uterine leiomyomas were used as Controls. The selection of the control group, and thus the whole study design, was then challenged by some reviewers, as regrettably, the correlation between this SNP and uterine fibroids had not been established at that point. Encouraged by that and to elucidate whether this DNA marker has any influence on uterine leiomyomas, we have decided to go a step further and examine disease-free controls. Until now, to our best knowledge ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) still has not been analyzed in uterine leiomyomas. The aim of this study was to analyze the frequencies of alleles and genotypes of Single Nucleotide Polymorphism ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) and to correlate it with the risk of uterine fibroids.
Study Design
Patients
The Test Group comprised 90 women treated surgically for UFs in the Department of Operative Gynecology, Endoscopy and Gynecologic Oncology, Polish Mother’s Memorial Hospital-Research Institute, Lodz, Poland. 90 healthy age- and BMI-matched individuals were used as Controls. DNA of the latter was provided by BioBank Laboratory (University of Lodz, Poland). For further statistical investigation of potential role of SNP
ADIPOQ(NM_004797.4):c.214+62G>T (rs1501299) in obesity and UFs, both groups have been stratified accordingly to total body fat content (using BMI—Body Mass Index—as a marker), into lean (BMI < 25), overweight (25 ≤ BMI < 30) and obese (BMI ≥ 30) and thus six groups (30 patients each) were obtained.
Test Group 1 (TG1)—BMI < 25 (n = 30)
Test Group 2 (TG2)—25 ≤ BMI < 30 (n = 30)
Test Group 3 (TG3)—BMI ≥ 30 (n = 30)
Controls 1 (C1)—BMI < 25 (n = 30)
Controls 2 (C2)—BMI 25 ≤ BMI < 30 (n = 30)
Controls 3 (C3)—BMI ≥ 30 (n = 30)
The summaries of both Test Group and Controls are presented in Tables 1, 2.
TABLE 1
| Age: mean (median, SD) | BMI: mean (median, SD) | |
|---|---|---|
| TG1 | 58.6 (54; ± 12.5) | 22.7 kg/m2 (23.3 kg/m2; ± 1.6) |
| TG2 | 60.9 (57.5; ± 11.2) | 27.9 kg/m2 (28.1 kg/m2; ± 1.3 ) |
| TG3 | 63.8 (64; ± 9.5) | 34.9 kg/m2 (34.5 kg/m2; ± 1.9) |
| In total | 61.1 (62; ± 11.2) | 28.6 kg/m2 (28.1 kg/m2; ± 5.2) |
Test group.
TABLE 2
| Age: mean (median; SD) | BMI: mean (median; SD) | |
|---|---|---|
| C1 | 54.3 (54; ± 4.2) | 22.6 kg/m2 (23.1 kg/m2; ± 1.9) |
| C2 | 57.5 (56; ± 4.6) | 27.2 kg/m2 (27.5 kg/m2; ± 0.2) |
| C3 | 57.5 (55; ± 6.2) | 33.4 kg/m2 (33.2 kg/m2; ± 2.5) |
| In total | 56.4 (55; ± 5.3) | 27.9 kg/m2 (27.5 kg/m2; ± 5.0) |
Controls.
Due to a well-established role of the investigated SNP in metabolic disorders and its potential significance in cancer development, a history of any such comorbidity was an exclusion criterion of the study. The study received internal funding grant from Polish Mother’s Memorial Hospital-Research Institute, Lodz, Poland (grant no. 2015/VII/29-MN).
Genotype Determination
To investigate the Test Group, DNA was retrieved from archival postoperative specimens (paraffin blocks stored in the Department of Clinical Pathology, Polish Mother’s Memorial Hospital-Research Institute, Lodz, Poland). Tissue samples, after original fixation in formaldehyde and embedding in paraffin, were microtome-sectioned at thicknesses of 5 µm and stained with hematoxylin and eosin. Then, the slices were placed in Eppendorf
®micro test tubes, shaken five times with xylene which was followed by 3-minute-long centrifugation (14,000 RPM) after each shaking. The sediment was lavaged in 96% ethanol, again centrifuged for 3 min and then dried in 37°C. DNA was extracted from the material by DNeasy Blood & Tissue Kit (Qiagen, Germany) according to manufacturer’s manual. DNA samples of Controls were provided by BioBank Laboratory (University of Lodz, Poland). For both Test Group and Controls, Polymerase Chain Reaction - Restriction Fragment Length Polymorphism (PCR-RFLP) was used to determine the genotypes of SNP
ADIPOQ(NM_004797.4):c.214+62G>T (rs1501299). Following primers were applied (Polgen, Poland):
Forward: 5′TCTCTCCATGGCTGACAGTG3′
Reverse: 5′AGATGCAGCAAAGCCAAAGT3′
The PCR-RFLP was completed in PTC-100 TM unit (MJ Research, INC, Waltham, MA, United States). The amplification was performed in 50 μl of reaction mixture which consisted of: genomic DNA, PCR buffer (TaKaRa, Japan), dNTP (TaKaRa, Japan), Taq Polymerase (TaKaRa, Japan), primers (Polgen, Poland) and deionized H2O. PCR cycler conditions were as follows: 95°C for 30 s, 62°C for 30 s and 72°C for 30 s—repeated for 35 cycles. The product (set in 20 μl of reaction mixture) was incubated in 65°C for 14 h with restriction enzyme (BsmI, New England BioLabs Inc., United States). PCR-RFLP products were then electrophoresed in a 2% agarose gel (Sigma, Saint Louis, United States) and visualized by ethidium bromide staining (Sigma, Saint Louis, United States). DNA Ladder 100 bp (Polgen, Poland) was used as mass ruler. Agarose gel was studied in ultraviolet light (Kodak Edas 290). The reaction produced fragments of 468bp (homozygous: GG), 468, 320 and 148 bp (heterozygous: GT) and 320 and 148 bp (homozygous: TT).
Statistical Analysis
χ2-test was used to assess the departure from Hardy-Weinberg equilibrium. Genotype and allele frequencies in Test Group and Controls were compared by χ2-test. Specific risks were expressed as odds ratios (ORs) with associated 95% confidence intervals (CIs) and adjusted to the logistic regression model. p-Values < 0.05 were considered significant.
Results
Table 3 displays the detailed distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in Test Group and in Controls with a further subdivision accordingly to BMI. The statistical analysis did not reveal significant differences in the distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) between Test Group and Controls—see Table 4. Graphic illustration is provided in Figure 1.
TABLE 3
| Test group (n = 90) | Controls (n = 90) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TG1 (n = 30) | TG2 (n = 30) | TG3 (n = 30) | C1 (n = 30) | C2 (n = 30) | C3 (n = 30) | |||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Genotype | ||||||||||||
| GG | 5 | 17 | 12 | 40 | 11 | 37 | 11 | 37 | 12 | 40 | 10 | 33 |
| GT | 19 | 63 | 14 | 47 | 15 | 50 | 16 | 53 | 11 | 37 | 16 | 53 |
| TT | 6 | 20 | 4 | 13 | 4 | 13 | 3 | 10 | 7 | 23 | 4 | 13 |
| Total | 30 | 100 | 30 | 100 | 30 | 100 | 30 | 100 | 30 | 100 | 30 | 100 |
| Allele | ||||||||||||
| G | 29 | 48 | 38 | 63 | 37 | 62 | 38 | 63 | 35 | 58 | 36 | 60 |
| T | 31 | 52 | 22 | 37 | 23 | 38 | 22 | 37 | 25 | 42 | 24 | 40 |
| Total | 60 | 100 | 60 | 100 | 60 | 100 | 60 | 100 | 60 | 100 | 60 | 100 |
Detailed distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in Test Group and in Controls.
TABLE 4
| Genotype/Allele | Test group (n = 90) | Controls (n = 90) | OR (95% PU)a | p b | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| GG | 28 | 31 | 33 | 37 | 1.00 Ref. c | |
| GT | 48 | 53 | 43 | 48 | 0.76 [0.39–1.45] | 0.507 |
| TT | 14 | 16 | 14 | 16 | 0.84 [0.34–2.07] | 0.887 |
| G | 104 | 58 | 109 | 61 | 1.00 Ref. c | |
| T | 76 | 42 | 71 | 39 | 0.89 [0.59–1.35] | 0.671 |
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in Test Group and in Controls.
Odds ratio analysis [OR, odds ratio; CI, confidence interval 95%].
For the dparture from Hardy-Weinberg equilibrium.
Reference wild allele.
FIGURE 1
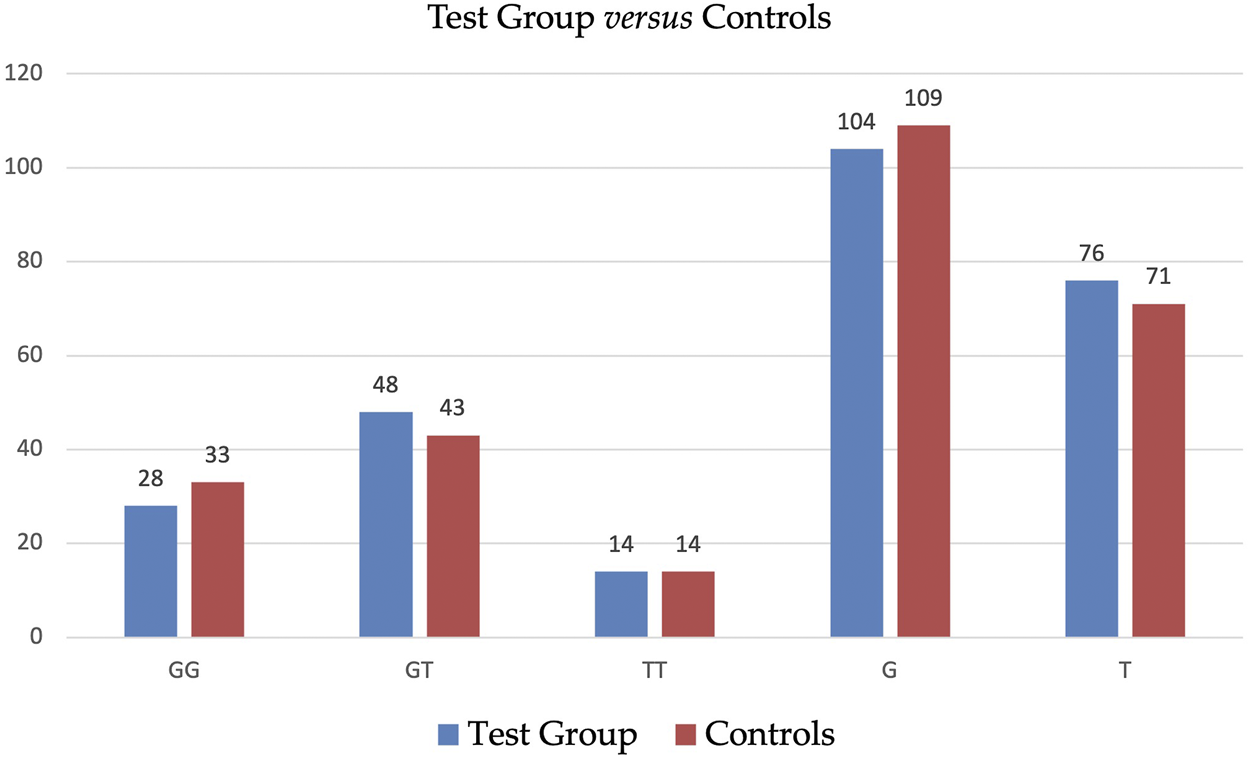
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in Test Group and in Controls.
Similarly, BMI-adjusted analysis of subgroups (i.e., TG1 vs. C1, TG2 vs. C2, TG3 vs. C3) has not revealed any statistically significant outcomes. However, the analysis revealed that allele G in TG3 is significantly more frequent (67 vs. 48%), and allele T in these patients is significantly less frequent (33 vs. 52%) than in C1 (see Table 5 and Figure 2).
TABLE 5
| Genotype/Allele | TG3 (n = 30) | C1 (n = 30) | OR (95% CI)a | p b | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| GG | 12 | 40 | 5 | 17 | 1.00 Ref.c | |
| GT | 16 | 53 | 19 | 63 | 0.35 [0.10–1.21] | 0.163 |
| TT | 2 | 7 | 6 | 20 | 0.14 [0.02–0.94] | 0.043 |
| G | 40 | 67 | 29 | 48 | 1.00 Ref.c | |
| T | 20 | 33 | 31 | 52 | 0.47 [0.22–0.97] | 0.042 |
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in TG3 vs. C1.
Odds ratio analysis [OR, odds ratio; CI, Confidence Interval 95%].
χ2 for the departure from Hardy-Weinberg equilibrium.
Reference: wild allele.
FIGURE 2
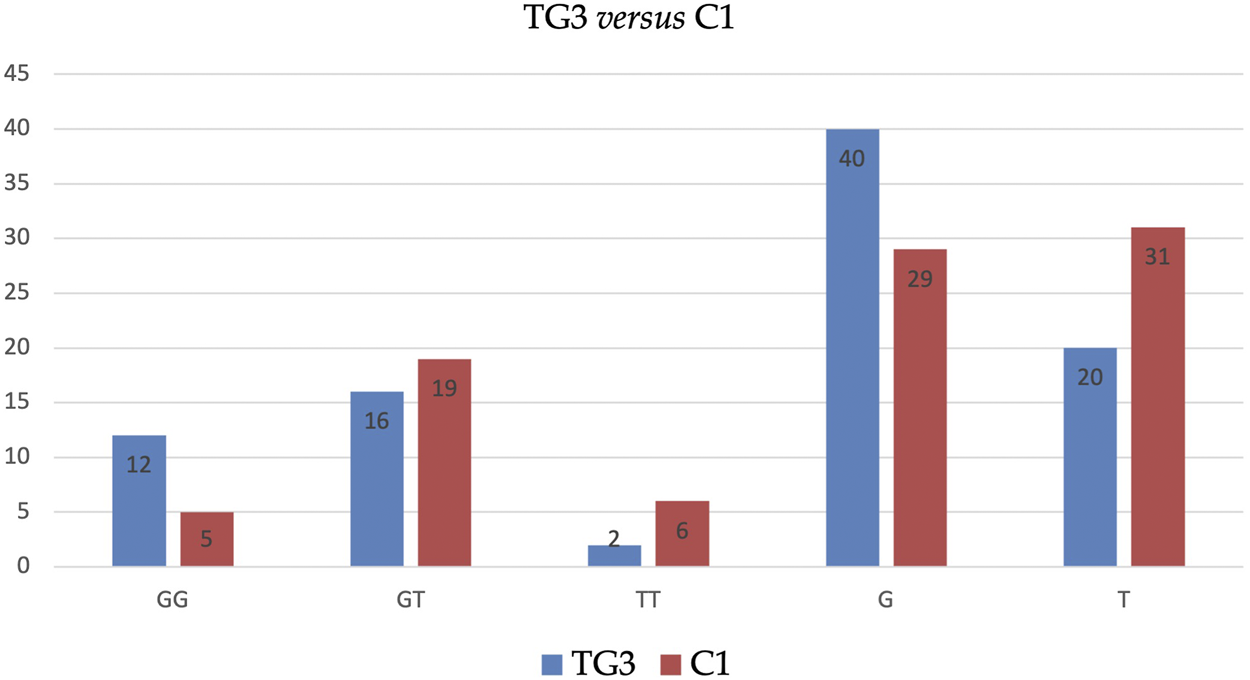
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in TG3 vs. C1.
In further analysis, correlations within the main groups were investigated: lean women with UFs (TG1) were tested versus obese ones (TG3) and lean controls (C1) versus obese controls (C3). Alike, no statistically significant findings were observed within the two abovementioned two comparisons (see Tables 6, 7).
TABLE 6
| Genotype/Allele | Lean (TG1) (n = 30) | Obese (TG3) (n = 30) | OR (95% CI)a | p b | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| GG | 5 | 17 | 11 | 37 | 1.00 Ref.c | |
| GT | 19 | 63 | 15 | 50 | 2.78 [0.79–9.78] | 0.186 |
| TT | 6 | 20 | 4 | 13 | 3.30 [0.63–17.16] | 0.150 |
| G | 29 | 48 | 37 | 62 | 1.00 Ref.c | |
| T | 31 | 52 | 23 | 38 | 1.71 [0.83–3.55] | 0.198 |
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) within Test Group: lean (TG1, BMI < 25) vs. obese (TG3, BMI ≥ 30).
Odds ratio analysis [OR, odds ratio; CI, Confidence Interval 95%].
χ2 for the departure from Hardy-Weinberg equilibrium.
Reference: wild allele.
TABLE 7
| Genotype/Allele | Lean (C1) (n = 30) | Obese (C3) (n = 30) | OR (95% CI)a | p b | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| GG | 11 | 37 | 10 | 33 | 1.00 Ref.c | |
| GT | 16 | 53 | 16 | 53 | 0.91 [0.30–2.73] | 0.920 |
| TT | 3 | 10 | 4 | 13 | 0.68 [0.121–3.82] | 0.499 |
| G | 38 | 63 | 36 | 60 | 1.00 Ref.c | |
| T | 22 | 37 | 24 | 40 | 0.86 [0.41–1.81] | 0.841 |
Distribution of genotypes and alleles of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) within Controls: lean (C1, BMI < 25) vs. obese (C3, BMI ≥ 30).
Odds ratio analysis [OR, odds ratio; CI, Confidence Interval 95%].
χ2 for the departure from Hardy-Weinberg equilibrium.
Reference: wild allele.
Discussion
As mentioned in the Introduction, this research was partly motivated by our previous study [31] and some concerns raised then over the selection of controls. By that time, and until this day, the role of SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) in UFs has not been elucidated. To explore this issue, in the current study we enrolled some new samples from disease-free age- and BMI-matched individuals and conducted a Pilot Study on this issue. This study extension was enabled by the newly established cooperation between our Institution and BioBank (University of Lodz, Poland), which provided us with suitable disease-free samples. This Pilot Study revealed no statistically significant differences in the distribution of genotypes and alleles of Single Nucleotide Polymorphism ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) between the main groups, which allows to draw a preliminary conclusion, that uterine leiomyomas are not correlated with this SNP (see Table 4). However, a statistically significant correlation was observed when obese Test Group patients were juxtaposed with lean Controls (TG3 vs. C1), where allele G proved to be more frequent, while allele T less frequent in the former group (Table 5 and Figure 2). It is noteworthy, that this result concerns alleles alone with no impact on the distribution of complete genotypes. Furthermore, this association is rather weak, and no definite conclusions should be drawn basing on this finding only. Besides, this weak association can be easily challenged, as it was solely observed between groups that differ not only by uterine fibroids’ presence, but—more importantly—by total body fat content (expressed by BMI) which may play a role in the distribution of genotypes and alleles of the studied SNP [21–26]. Interestingly, in our research the distribution of genotypes and alleles of the studied SNP was not associated with BMI (Tables 6, 7) which rather suggests that the observed weak correlation should not be attributed to overall body adiposity.
Moreover, one must mention the limitations of our research. To begin with, the sample number is low and may be considered quantitively unsatisfactory to make final conclusions in a genetic study [32, 33]. Furthermore, an issue of studying a single SNP may also be raised as an obvious drawback here. Therefore, taking into consideration the preliminary data of our pilot study, we believe that further research in this field is needed.
Conclusion
According to our Pilot Study, SNP ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) is not associated with uterine leiomyomas. Taking into consideration some clear limitations of our research, conclusions should be drawn with caution. Further research on larger groups is warranted to improve the credibility of our finding.
Statements
Data availability statement
The raw data supporting the conclusions of this article is stored at the Institution and will be made available by the authors upon request.
Ethics statement
The studies involving human participants were reviewed and approved by The Local Ethic Committee by Polish Mother’s Memorial Hospital-Research Institute. The patients/participants provided their written informed consent to participate in this study.
Author contributions
JB: protocol and project development, data collection and management, manuscript writing and editing. AS: manuscript writing and editing. MW, GJ, and AO: data collection and management. BS and HR: genetical assays and data analysis, scientific supervision. AM: protocol and project development, scientific supervision.
Funding
Internal grant of Polish Mother’s Memorial Hospital-Research Institute no. 2015/VII/29-MN.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Commandeur A. E. Styer A. K. Teixeira J. M . Epidemiological and Genetic Clues for Molecular Mechanisms Involved in Uterine Leiomyoma Development and Growth. Hum Reprod Update (2015) 21(5):593–615. 10.1093/humupd/dmv030
2.
Giuliani E. As Sanie S. Marsh E. E . Epidemiology and Management of Uterine Fibroids. Int J Gynecol Obstet (2020) 149(1):3–9. Epub 2020 Feb 17. 10.1002/ijgo.13102
3.
Gurusamy K. S. Vaughan J. Fraser I. S. Best L. M. J. Richards T. Medical Therapies for Uterine Fibroids - A Systematic Review and Network Meta-Analysis of Randomised Controlled Trials. PLoS One (2016) 11(2):e0149631. 10.1371/journal.pone.0149631
4.
Ayyad I . Relationship between Fibroid Uterus and Obesity. MEJFM (2011) 9(7):p7–9. 3p. 10.5742/mejfm.2011.97071
5.
Shikora S. A. Niloff J. M. Bistrian B. R. Forse R. A. Blackburn G. L . Relationship between Obesity and Uterine Leiomyomata. Nutrition (1991) 7(4):251–5.
6.
Välimäki N. Kuisma H. Pasanen A. Heikinheimo O. Sjöberg J. Bützow R. et al Genetic Predisposition to Uterine Leiomyoma Is Determined by Loci for Genitourinary Development and Genome Stability. Elife (2018) 7:e37110. Published 2018 Sep 18. 10.7554/eLife.37110
7.
Moravek M. B. Bulun S. E. Endocrinology of Uterine Fibroids. Curr Opin Obstet Gynecol (2015) 27(4):276–83. 10.1097/gco.0000000000000185
8.
Simón C . Introduction: Are We Advancing in Our Scientific Understanding and Therapeutic Improvement of Uterine Fibroids or Not. Fertil Steril (2014) 102(3):611–2. 10.1016/j.fertnstert.2014.07.1210
9.
Mehine M. Mäkinen N. Heinonen H.-R. Aaltonen L. A. Vahteristo P . Genomics of Uterine Leiomyomas: Insights from High-Throughput Sequencing. Fertil Sterility (2014) 102(3):621–9. 10.1016/j.fertnstert.2014.06.050
10.
Giralt M. Cereijo R. Villarroya F . Adipokines and the Endocrine Role of Adipose Tissues. Handb Exp Pharmacol (2016) 233:265–82. 10.1007/164_2015_6
11.
Tilg H Moschen A. R. Adipocytokines: Mediators Linking Adipose Tissue, Inflammation and Immunity. Nat Rev Immunol (2006) 6(10):772–83. 10.1038/nri1937
12.
Arita Y. Kihara S. Ouchi N. Takahashi M. Maeda K. Miyagawa J.-I. et al Paradoxical Decrease of an Adipose-specific Protein, Adiponectin, in Obesity. Biochem Biophysical Res Commun (1999) 257(1):79–83. 10.1006/bbrc.1999.0255
13.
Hotta K. Funahashi T. Bodkin N. L. Ortmeyer H. K. Arita Y. Hansen B. C et al Circulating Concentrations of the Adipocyte Protein Adiponectin Are Decreased in Parallel with Reduced Insulin Sensitivity during the Progression to Type 2 Diabetes in Rhesus Monkeys. Diabetes (2001) 50(5):1126–33. 10.2337/diabetes.50.5.1126
14.
Yamauchi T. Kamon J. Waki H. Terauchi Y. Kubota N. Hara K. et al The Fat-Derived Hormone Adiponectin Reverses Insulin Resistance Associated with Both Lipoatrophy and Obesity. Nat Med (2001) 7(8):941–6. 10.1038/90984
15.
Soliman P. T. Wu D. Tortolero-Luna G. Schmeler K. M. Slomovitz B. M. Bray M. S. et al Association between Adiponectin, Insulin Resistance, and Endometrial Cancer. Cancer (2006) 106(11):2376–81. 10.1002/cncr.21866
16.
Petridou E. Mantzoros C. Dessypris N. Koukoulomatis P. Addy C. Voulgaris Z et al Plasma Adiponectin Concentrations in Relation to Endometrial Cancer: a Case-Control Study in Greece. J Clin Endocrinol Metab (2003) 88(3):993–7. 10.1210/jc.2002-021209
17.
Rzepka-Górska I. Bedner R. Cymbaluk-Płoska A. Chudecka-Głaz A. Serum Adiponectin in Relation to Endometrial Cancer and Endometrial Hyperplasia with Atypia in Obese Women. Eur J Gynaecol Oncol (2008) 29(6):594–7.
18.
Cust A. E. Kaaks R. Friedenreich C. Bonnet F. Laville M. Lukanova A. et al Plasma Adiponectin Levels and Endometrial Cancer Risk in Pre- and Postmenopausal Women. J Clin Endocrinol Metab (2007) 92(1):255–63. Epub 2006 Oct 24. 10.1210/jc.2006-1371
19.
Ma Y. Liu Z. Zhang Y. Lu B. Serum Leptin, Adiponectin and Endometrial Cancer Risk in Chinese Women. J Gynecol Oncol (2013) 24(4):336–41. 10.3802/jgo.2013.24.4.336
20.
Melistas L. Mantzoros C. S. Kontogianni M. Antonopoulou S. Ordovas J. M. Yiannakouris N. Association of the +45T>G and +276G>T Polymorphisms in the Adiponectin Gene with Insulin Resistance in Nondiabetic Greek Women. Eur J Endocrinol (2009) 161(6):845–52. Epub 2009 Sep 15. PMID: 19755407; PMCID: PMC2896503. 10.1530/EJE-09-0492
21.
Ukkola O. Ravussin E. Jacobson P. Sjöström L. Bouchard C . Mutations in the Adiponectin Gene in Lean and Obese Subjects from the Swedish Obese Subjects Cohort. Metabolism (2003) 52(7):881–4. PMID: 12870165. 10.1016/s0026-0495(03)00074-x
22.
Salmenniemi U. Zacharova J. Ruotsalainen E. Vauhkonen I. Pihlajamäki J. Kainulainen S. et al Association of Adiponectin Level and Variants in the Adiponectin Gene with Glucose Metabolism, Energy Expenditure, and Cytokines in Offspring of Type 2 Diabetic Patients. J Clin Endocrinol Metab (2005) 90(7):4216–23. Epub 2005 Apr 26. PMID: 15855264. 10.1210/jc.2004-2289
23.
Zayani N. Omezzine A. Boumaiza I. Achour O. Rebhi L. Rejeb J. et al Association of ADIPOQ, Leptin, LEPR, and Resistin Polymorphisms with Obesity Parameters in Hammam Sousse Sahloul Heart Study. J Clin Lab Anal (2017) 31(6):e22148. Epub 2017 Feb 13. 10.1002/jcla.22148
24.
Christodoulou A. Ierodiakonou D. Awofala A. A. Petrou M. Kales S. N. Christiani D. C. et al Variants in ADIPOQ Gene Are Linked to Adiponectin Levels and Lung Function in Young Males Independent of Obesity. PLoS One (2020) 15(1):e0225662. 10.1371/journal.pone.0225662
25.
Kaur H. Badaruddoza B. Bains V. Kaur A. Genetic Association of ADIPOQ Gene Variants (-3971A>G and +276G>T) with Obesity and Metabolic Syndrome in North Indian Punjabi Population. PLoS One (2018) 13(9):e0204502. 10.1371/journal.pone.0204502
26.
Filippi E. Sentinelli F. Trischitta V. Romeo S. Arca M. Leonetti F. et al Association of the Human Adiponectin Gene and Insulin Resistance. Eur J Hum Genet (2004) 12(3):199–205. 10.1038/sj.ejhg.5201120
27.
Xita N. Georgiou I. Chatzikyriakidou A. Vounatsou M. Papassotiriou G.-P. Papassotiriou I. et al Effect of Adiponectin Gene Polymorphisms on Circulating Adiponectin and Insulin Resistance Indexes in Women with Polycystic Ovary Syndrome. Clin Chem (2005) 51(2):416–23. Epub 2004 Dec 8. PMID: 15590747. 10.1373/clinchem.2004.043109
28.
Warodomwichit D. Shen J. Arnett D. K. Tsai M. Y. Kabagambe E. K. Peacock J. M. et al ADIPOQ Polymorphisms, Monounsaturated Fatty Acids, and Obesity Risk: the GOLDN Study. Obesity (Silver Spring) (2009) 17(3):510–7. 10.1038/oby.2008.583
29.
National Center For Biotechnology Information (2021). Short Genetic Variations. Available from: https://www.ncbi.nlm.nih.gov/snp/rs1501299.
30.
Cheung C. Y. Y. Hui E. Y. L. Cheung B. M. Y. Woo Y. C. Xu A. Fong C. H. Y. et al Adiponectin Gene Variants and the Risk of Coronary Heart Disease: a 16-year Longitudinal Study. Eur J Endocrinol (2014) 171(1):107–15. Epub 2014 Apr 23. PMID: 24760538. 10.1530/EJE-14-0079
31.
Bieńkiewicz J. Smolarz B. Malinowski A. Association Between Single Nucleotide Polymorphism +276G > T (Rs1501299) in ADIPOQ and Endometrial Cancer. Pathol Oncol Res (2016) 22(1):135–8. Epub 2015 Sep 19. 10.1007/s12253-015-9985-9
32.
Hong E. P. Park J. W. Sample Size and Statistical Power Calculation in Genetic Association Studies. Genomics Inform (2012) 10(2):117–22. Epub 2012 Jun 30. 10.5808/GI.2012.10.2.117
33.
B-Rao C . Sample Size Considerations in Genetic Polymorphism Studies. Hum Hered (2001) 52(4):191–200. 10.1159/000053376
Summary
Keywords
single nucleotide polymorphism, obesity, uterine leiomyomas, uterine fibroids, c.276 G>T, adiponectin, ADIPOQ, (NM_004797.4):c.214+62G>T
Citation
Bieńkiewicz J, Smolarz B, Wilczyński M, Stepowicz A, Jabłoński G, Obłękowska A, Malinowski A and Romanowicz H (2022) Is Single Nucleotide Polymorphism ADIPOQ (NM_004797.4):c.214+62G>T (rs1501299) Associated With Uterine Leiomyomas? A Pilot Study. Pathol. Oncol. Res. 27:1609966. doi: 10.3389/pore.2021.1609966
Received
18 July 2021
Accepted
29 December 2021
Published
18 February 2022
Volume
27 - 2022
Edited by
József Tímár, Semmelweis University, Hungary
Updates
Copyright
© 2022 Bieńkiewicz, Smolarz, Wilczyński, Stepowicz, Jabłoński, Obłękowska, Malinowski and Romanowicz.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Bieńkiewicz, jan.bienkiewicz@iczmp.edu.pl
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.