- 1Faculty of Medicine, Vilnius University, Vilnius, Lithuania
- 2Hematology, Oncology and Transfusion Medicine Center, Vilnius University Hospital Santaros Klinikos, Vilnius, Lithuania
- 3Department of Experimental, Preventive and Clinical Medicine, State Research Institute Centre for Innovative Medicine, Vilnius, Lithuania
- 4Center for Pediatric Oncology and Hematology, Vilnius University Hospital Santaros Klinikos, Vilnius, Lithuania
Acute lymphoblastic leukemia (ALL) with recurrent genetic lesions, affecting a series of kinase genes, is associated with unfavorable prognosis, however, it could benefit from treatment with tyrosine kinase inhibitors (TKI). NUP214::ABL1 fusion is detected in 6% of T-cell acute lymphoblastic leukemia (T-ALL), and is very rare in B-ALL. We present a case of adolescent with B-ALL and a cryptic NUP214::ABL1 fusion which was initially missed during diagnostic screening and was detected by additional RNA sequencing. Treatment with specific ABL-inhibitor Imatinib was added later in therapy with a good effect. Initial treatment according to conventional chemotherapy was complicated by severe side effects. At the end of Consolidation, the patient was stratified to a high risk group with allogeneic hematopoietic stem cell transplantation because of insufficient response to therapy. At that time, targeted RNA sequencing detected NUP214::ABL1 gene fusion which was previously missed due to a small microduplication in the 9q34 chromosome region. Gene variant analysis revealed no TKI-resistant ABL1 mutations; therefore, treatment with Imatinib was added to target the NUP214::ABL1 fusion protein. A negative minimal residual disease was achieved, and treatment was downgraded to intermediate risk protocol. Combining routine genetic assays with next-generation sequencing methods could prevent from missing atypical gene alterations. Identification of rare targetable genetic subtypes is of importance in order to introduce targeted therapy as early as possible that may improve survival and reduce toxicity. Treatment with ABL1 inhibitor imatinib mesylate revealed as a highly effective targeted therapy against the leukemia driving protein kinase.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease with certain cytogenetic aberrations being long-recognized to be anticipated an unfavourable prognosis. A new BCR-ABL-like subgroup of tyrosine-kinase driven ALL has been associated with a poor response to chemotherapy, a high relapse risk, and unfavorable long-term outcomes (1). In the 2016 updated World Health Organization classification of myeloid neoplasms and acute leukemia, BCR-ABL1-like B-ALL was added as a new provisional entity (2). Its gene expression profile is similar to BCR::ABL1, however, is lacking BCR::ABL1 fusion (3). The presence of Nucleoporin 214-ABL Proto-Oncogene 1 (NUP214::ABL1) gene fusion is detected in 6% of T-ALL, whereas it is rare in B-ALL (4). A series of genes that activate tyrosine kinase and cytokine receptor signaling are affected in BCR-ABL1-like ALL, suggesting the potential interest of targeted treatment with tyrosine kinase inhibitors (TKI) (5). However, the therapeutic effect of TKI for the NUP214::ABL1-positive patients is disputable as clinical experience is limited (6–11). Standard worldwide screening methods for known ABL1 gene fusions include fluorescence in situ hybridization (FISH) analysis and reverse transcription polymerase chain reaction (RT-PCR) (12). Nevertheless, these screening techniques detect a limited number of alterations as BCR-ABL1-like ALL is known for its highly heterogeneous background (13).
We present a case of pediatric B-ALL with a cryptic NUP214::ABL1 gene fusion which was initially missed during diagnostic screening due to unusual genetic alteration and was identified by the targeted next-generation sequencing (NGS) only. Treatment with a first-generation TKI (imatinib) was added to the chemotherapy with a good effect.
Case Description
A 15 year-old boy with no previous significant medical history was admitted to our pediatric department in July 2020 for high fever, petechial and hemorrhagic rash, and vomiting. The blood count showed hemoglobin 51 g/L, platelet count 34 × 109/L, and hyperleukocytosis 464.5 109/L. Immunophenotyping confirmed the expression of B-lymphoid markers CD45, CD19, CD10, CD20, CD81, CD22, cCD22, CD24, and cCD79a. Routine genomic screening by a single nucleotide polymorphism array (SNP-A) detected normal male karyotype 46,XY without larger aberrations in size ≥5 Mb. FISH and RT-PCR did not detect any of the following recurrent rearrangements: BCR::ABL1, KMT2A, EPOR, ABL1, ABL2, RUNX1 (CSF1R), PDGFRB, E2A (TCF3), JAK2, ETV6::RUNX1, or CRLF2. Cerebrospinal fluid showed three WBC/μl and ∼5.8% of aberrant phenotype B-lymphoid cells, with no leukemic blasts in cytospin. B-ALL, CNS1 was diagnosed.
Treatment was conducted according to ALLTogether protocol Induction B with dexamethasone, vincristine, daunorubicin, pegylated-asparaginase (PEG-Asp), and intrathecal methotrexate. At the end of induction, at time point 1 (TP1), minimal residual disease (MRD) in bone marrow showed residual cells of 0.79% by flow cytometry (FC) and 0.03% by IG/TCR quantitative PCR, respectively. Discrepancy in the lab results was interpreted as a subclone of leukemic cells that was not captured by PCR, and the patient was stratified to intermediate-high risk (IR-H) due to a slow response to the therapy as per protocol (Figure 1). Consolidation with dexamethasone, vincristine, 6-mercaptopurine, cyclophosphamide, cytarabine, PEG-Asp, and intrathecal methotrexate was given according to IR-H protocol. Bone marrow evaluation on day 71, time point 2 (TP2) still showed positive MRD: ∼0.18% and 0.07% by FC and IG/TCR quantitative PCR respectively. The patient was stratified into High-Risk (HR) group with allogeneic hematopoietic stem cell transplantation (allo-HSCT) (Figure 1).
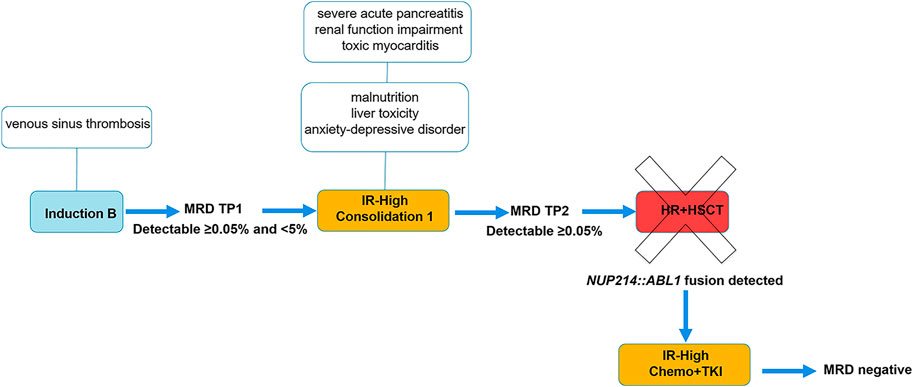
FIGURE 1. Treatment flowchart and timeline of symptoms. Abbreviations: HR+HSCT, high risk block chemotherapy with hematopoietic stem cell transplantation; IR-High, intermediate-high risk group; MRD, minimal residual disease; TKI, tyrosine kinase inhibitor; TP1, time point 1: end of Induction, day 29; TP2, time point 2: end of Consolidation 1, day 71.
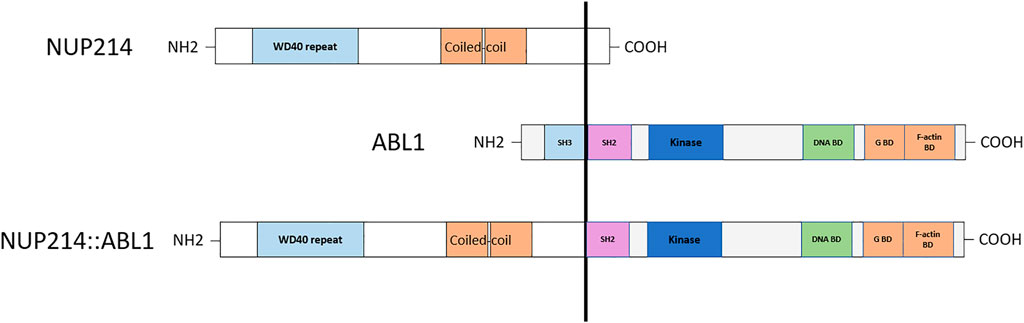
Chemotherapy was complicated by multiple side effects. The patient was treated at the intensive care unit twice because of repeated tonic-clonic seizures caused by venous sinus thrombosis in Induction phase and because of severe acute pancreatitis in Consolidation 1. Acute pancreatitis was complicated by multiple organ dysfunction including renal function impairment and toxic myocarditis. Furthermore, the patient suffered from malnutrition, liver toxicity and mixed anxiety-depressive disorder. On day 71 (TP2), targeted RNA sequencing was performed on patient’s RNA sample using TruSight Pan-Cancer sequencing kit as described earlier (14). Sequencing data analysis revealed t(9;9)(q34;q34)/NUP214::ABL1 gene fusion. Exon 33 of the NUP214 gene and exon 3 of the ABL1 gene were fused. RT-PCR method was used to confirm NUP214::ABL1 fusion transcript. Gene variant analysis showed no TKI-resistant ABL1 mutations; therefore, treatment with a first-generation TKI imatinib mesylate was added to the conventional chemotherapy. Complete remission (CR) was achieved within a month, and treatment was downgraded to intermediate-risk protocol (Figure 1). At the time of writing the manuscript, the patient is in the first CR 24 months from diagnosis.
The present report was conducted in accordance with the guidelines of the Declaration of Helsinki. Institutional ethical review board permission for a case report was obtained and a written informed consent was received from the patient and his parents.
Discussion
Cryptic NUP214::ABL1 fusion is a rare genetic entity carrying kinase activating alterations and making the patients candidates for TKI treatment. Although ABL1 gene rearrangements are most commonly detected in B-ALL, NUP214::ABL1 fusion transcript is mainly described in T-ALL patients (7–9,11,15), whereas in B-ALL its expression is described in individual cases only. In T-ALL, chimeric NUP214::ABL1 protein showed to be sensitive to TKI in preclinical and clinical studies (11), whereas in B-ALL the role of TKI still needs to be established. In pediatric population, approximately 10%–15% of B-ALL cases reveal a BCR::ABL1-like profile representing a biologically and clinically challenging group (3). ABL class tyrosine kinase fusion genes are expected to be clonal leukemia drivers and usually respond well to ABL class inhibitors imatinib or dasatinib. Imatinib is generally regarded as the safest of the TKIs, with no long-term irreversible side effects. Although many authors recommend second or third-generation TKIs to override the frequent ATP-binding site mutations (16,17), in our case imatinib showed a very good effect, as there was no evidence of ABL1 mutations.
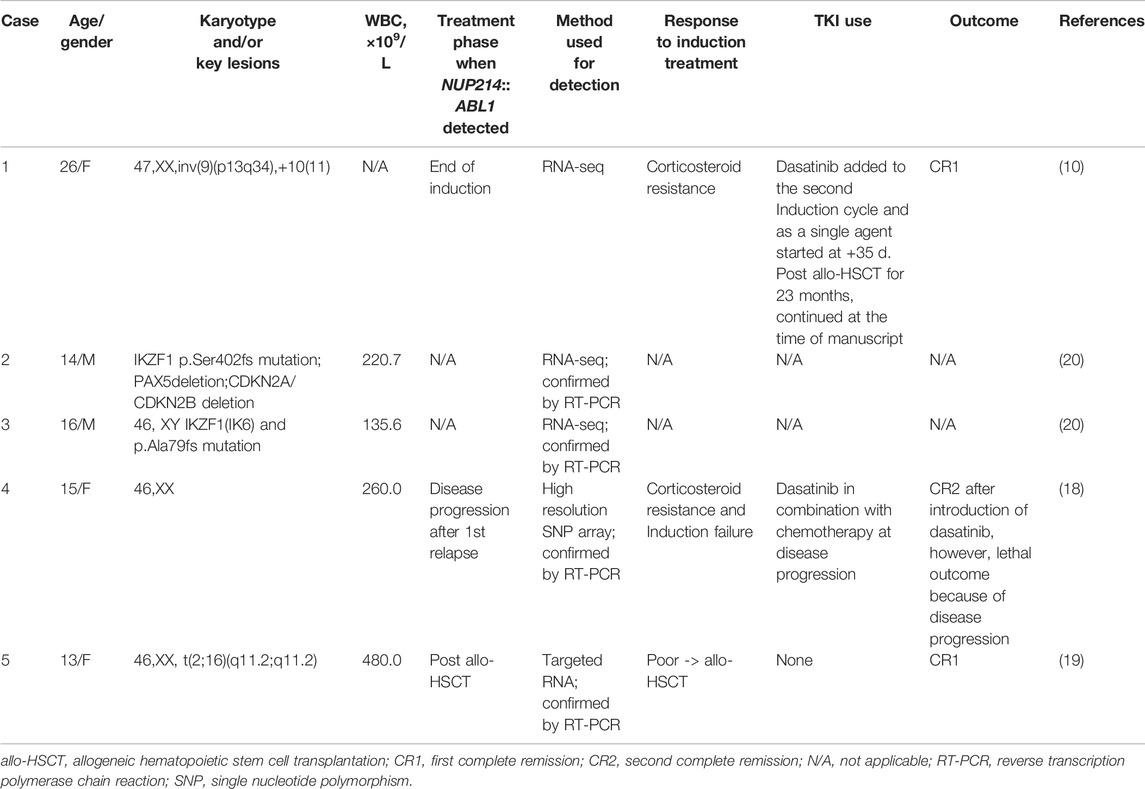
To the best of our knowledge, only five B-ALL cases with NUP214::ABL1 fusion caused by intrachromosomal microduplication had been published so far, data are summarized in Table 1. The patients reported were teenagers or young adults, all being >13 years old at the time of diagnosis. Four patients, for whom data was available, had high hyperleukocytosis (WBC >100 × 109/L) at presentation, similarly to our case. In all reported cases, the patients had a poor initial response to therapy and were stratified to very high risk (VHR) chemotherapy (case #4) or allogeneic hematopoietic stem cell transplantation (allo-HSCT) (cases #1 and #5). Two patients received treatment with TKI after allo-HSCT and achieved negative MRD, however, subsequent disease progression in case #4 resulted in lethal outcome. One patient (#5) underwent successful allo-HSCT without additional TKI use.
Unlike in previously reported cases, we initiated treatment with a first-generation TKI imatinib mesylate with high efficacy. Two cases (cases #1 and #4) reported a second-generation TKI dasatinib, and a choice of TKI was not specified in the cases #2 and #3. Mechanisms of resistance to imatinib are known to be related to the mutations of ATP-binding site in BCR::ABL1 positive ALL, therefore, dasatinib, nilotinib or ponatinib are preferred as first line therapy (16,17). In our case, gene variant analysis revealed no TKI resistant ABL1 mutations, which could explain a good effect of Imatinib which was added to the first line of conventional chemotherapy. This subsequently allowed to downgrade the treatment to IR-H risk thus evading allo-HSCT and potentially life-threatening further toxicity.
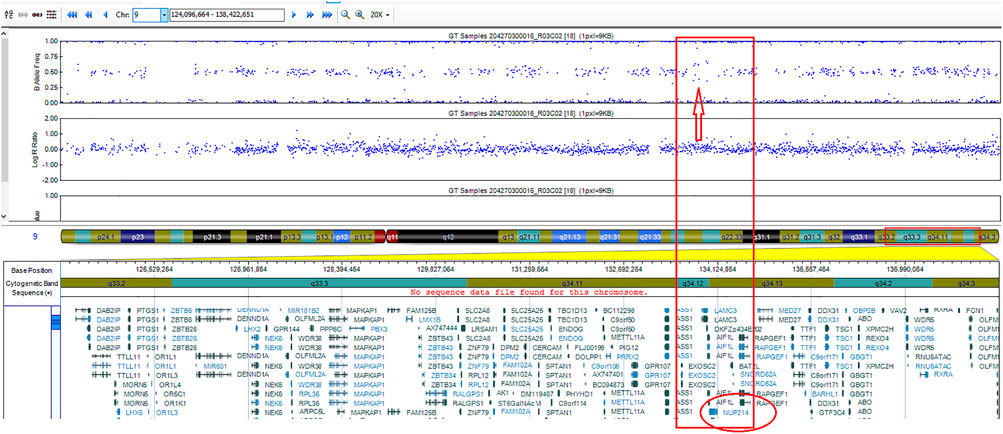
Detection of cryptic ABL1 gene rearrangements by conventional genetic analysis can be a challenge (20, 19). Among the reported cases, NUP214::ABL1 fusion was identified early in treatment, after the first cycle of induction, in one B-ALL case only, using NGS techniques (10). In two cases, the fusion was initially missed by routine diagnostic methods and detected later by SNP-A or targeted RNA sequencing (20, 19) (Table 1). In our case, the ABL1 gene break was initially missed by FISH array (ABL1 Break Apart Probe) due to a small 445 Kb microduplication in the 9q34 chromosome region and was detected later by performing targeted RNA sequencing. SNP-A karyotyping missed microduplication which was smaller than 5 Mb in size (Figure 2). NUP214 and ABL1 genes are located at the edges of the 9q34 region, therefore, FISH cannot successfully detect NUP214::ABL1 gene fusion due to technical limitations (Figure 3). This particular cryptic fusion mechanism was described in detail by Tsujimoto et al. (20). In our previous retrospective population-based BCR::ABL1-negative B-other ALL cohort study, we did not detect any ABL-class fusions in pediatric Lithuanian patients (14), making this case to be the only NUP214::ABL1 gene fusion case in Lithuanian childhood B-ALL emphasizing very rare incidence of this aberration. Some authors suggest that all patients with B-ALL should undergo NGS analysis in parallel with conventional genetic screening (13). In our case, adding TKI to the first line treatment enabled us to downgrade the treatment risk group for the patient. However, earlier NGS results detecting targetable genomic alteration would have been beneficial by allowing initiation of targeted therapy and possibly preventing severe drug-induced side effects.
Conclusion
Identification of rare targetable genetic subtypes is of importance in order to introduce individualized targeted therapy as early as possible to improve survival and reduce toxicity. Combining TKI with chemotherapy for ABL1 rearranged B-ALL should be considered for the first-line treatment. B-ALL in adolescent patients without detected recurrent cytogenetic or molecular abnormalities (B-others) should be immediately analyzed further by NGS methods to prevent from missing atypical gene alterations.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Vilnius University Hospital Santaros Klinikos. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
ES-R designed the study, collected, analyzed and interpreted the data and wrote the manuscript; GV designed and supervised the study, interpreted the data and critically reviewed the manuscript; RN and VD designed the study, performed genetic analysis, interpreted the data and critically reviewed the manuscript; SS designed the study, collected and interpreted the data and critically reviewed the manuscript. All authors agreed and approved the final version of manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ALL, acute lymphoblastic leukemia; allo-HSCT, allogeneic hematopoietic stem cell transplantation; CR, complete remission; FISH, fluorescence in situ hybridization; HR+HSCT, high risk block chemotherapy with hematopoietic stem cell transplantation; ICU, intensive care unit; IR-High, intermediate-high risk group; MRD, minimal residual disease; NGS, next-generation sequencing; PEG-Asp, pegylated-asparaginase; RT-PCR, reverse transcription polymerase chain reaction; SNP-A, single nucleotide polymorphism array; TKI, tyrosine kinase inhibitor; TP1, time point 1: end of Induction, day 29; TP2, time point 2: end of Consolidation 1, day 71; VHR, very high risk chemotherapy.
References
1. Tasian, SK, Loh, ML, and Hunger, SP. Philadelphia Chromosome-like Acute Lymphoblastic Leukemia. Blood (2017) 130(19):2064–72. doi:10.1182/blood-2017-06-743252
2. Arber, DA, Orazi, A, Hasserjian, R, Thiele, J, Borowitz, MJ, Le Beau, MM, et al. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood (2016) 127(20):2391–405. doi:10.1182/blood-2016-03-643544
3. Den Boer, ML, van Slegtenhorst, M, De Menezes, RX, Cheok, MH, Buijs-Gladdines, JGCAM, Peters, STCJM, et al. A Subtype of Childhood Acute Lymphoblastic Leukaemia with Poor Treatment Outcome: a Genome-wide Classification Study. Lancet Oncol (2009) 10(2):125–34. doi:10.1016/S1470-2045(08)70339-5
4. De Braekeleer, E, Douet-Guilbert, N, Rowe, D, Bown, N, Morel, F, Berthou, C, et al. ABL1 Fusion Genes in Hematological Malignancies: a Review. Eur J Haematol (2011) 86(5):361–71. doi:10.1111/j.1600-0609.2011.01586.x
5. Roberts, KG, Li, Y, Payne-Turner, D, Harvey, RC, Yang, YL, Pei, D, et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N Engl J Med (2014) 371:1005–15. doi:10.1056/NEJMoa1403088
6. Deenik, W, Beverloo, HB, van der Poel-van de Luytgaarde Scp, a. M, Wattel, MM, van Esser Jwj, , , Valk, PJM, et al. Rapid Complete Cytogenetic Remission after Upfront Dasatinib Monotherapy in a Patient with a NUP214-ABL1-Positive T-Cell Acute Lymphoblastic Leukemia. Leukemia (2009) 23(3):627–9. doi:10.1038/leu.2008.318
7. Stergianou, K, Fox, C, and Russell, NH. Fusion of NUP214 to ABL1 on Amplified Episomes in T-ALL-Iimplications for Treatment. Leukemia (2005) 19(9):1680–1. doi:10.1038/sj.leu.2403877
8. Koschmieder, S, Burmeister, T, Brüggemann, M, Berkemeier, A, Volpert, S, Wieacker, P, et al. Molecular Monitoring in NUP214-ABL-Positive T-Acute Lymphoblastic Leukemia Reveals Clonal Diversity and Helps to Guide Targeted Therapy. Leukemia (2014) 28(2):419–22. doi:10.1038/leu.2013.272
9. Tsurusaki, Y, Nagai, J, Fujita, S, Sugiyama, M, Nakamura, W, Hayashi, A, et al. Whole-exome Sequencing Reveals the Subclonal Expression of NUP214-ABL1 Fusion Gene in T-Cell Acute Lymphoblastic Leukemia. Pediatr Blood Cancer (2020) 67(1):e28019. doi:10.1002/pbc.28019
10. Aldoss, I, and Pullarkat, V. Response to Single Agent Dasatinib post Allogeneic Transplant in B-Cell Acute Lymphoblastic Leukemia with NUP214-ABL1. Leuk Lymphoma (2019) 60(11):2832–4. doi:10.1080/10428194.2019.1605510
11. Chen, Y, Zhang, L, Huang, J, Hong, X, Zhao, J, Wang, Z, et al. Dasatinib and Chemotherapy in a Patient with Early T-Cell Precursor Acute Lymphoblastic Leukemia and NUP214-ABL1 Fusion: A Case Report. Exp Ther Med (2017) 14(5):3979–84. doi:10.3892/etm.2017.5046
12. Coccaro, N, Anelli, L, Zagaria, A, Specchia, G, and Albano, F. Next-Generation Sequencing in Acute Lymphoblastic Leukemia. Int J Mol Sci (2019) 20(12):2929. doi:10.3390/ijms20122929
13. Sherali, N, Hamadneh, T, Aftab, S, Alfonso, M, and Tsouklidis, N. Integration of Next-Generation Sequencing in Diagnosing and Minimal Residual Disease Detection in Patients with Philadelphia Chromosome-like Acute Lymphoblastic Leukemia. Cureus (2020) 12(9):e10696. doi:10.7759/cureus.10696
14. Norvilas, R, Dirse, V, Semaskeviciene, R, Mickeviciute, O, Gineikiene, E, Stoskus, M, et al. Low Incidence of ABL-Class and JAK-STAT Signaling Pathway Alterations in Uniformly Treated Pediatric and Adult B-Cell Acute Lymphoblastic Leukemia Patients Using MRD Risk-Directed Approach - a Population-Based Study. BMC Cancer (2021) 21(1):326. doi:10.1186/s12885-020-07781-6
15. Burmeister, T, Gökbuget, N, Reinhardt, R, Rieder, H, Hoelzer, D, and Schwartz, S. NUP214-ABL1 in Adult T-ALL: the GMALL Study Group Experience. Blood (2006) 108(10):3556–9. doi:10.1182/blood-2006-04-014514
16. Rossari, F, Minutolo, F, and Orciuolo, E. Past, Present, and Future of Bcr-Abl Inhibitors: from Chemical Development to Clinical Efficacy. J Hematol Oncol (2018) 11:84. doi:10.1186/s13045-018-0624-2
17. Hunger, SP. Tyrosine Kinase Inhibitor Use in Pediatric Philadelphia Chromosome–Positive Acute Lymphoblastic Anemia. Hematol Am Soc Hematol Educ Program (2011) 2011(1):361–5. doi:10.1182/asheducation-2011.1.361
18. Roberts, KG, Morin, RD, Zhang, J, Hirst, M, Zhao, Y, Su, X, et al. Genetic Alterations Activating Kinase and Cytokine Receptor Signaling in High-Risk Acute Lymphoblastic Leukemia. Cancer Cell (2012) 22(2):153–66. doi:10.1016/j.ccr.2012.06.005
19. Duployez, N, Grzych, G, Ducourneau, B, Fuentes, MA, Grardel, N, Boyer, T, et al. NUP214-ABL1 Fusion Defines a Rare Subtype of B-Cell Precursor Acute Lymphoblastic Leukemia that Could Benefit from Tyrosine Kinase Inhibitors. Haematologica (2016) 101(4):e133–4. doi:10.3324/haematol.2015.136499
Keywords: case report, targeted therapy, tyrosine kinase inhibitors, acute lymphoblastic leukemia, imatinib, BCR-ABL1-like
Citation: Stukaite-Ruibiene E, Norvilas R, Dirse V, Stankeviciene S and Vaitkeviciene GE (2022) Case Report: Specific ABL-Inhibitor Imatinib Is an Effective Targeted Agent as the First Line Therapy to Treat B-Cell Acute Lymphoblastic Leukemia With a Cryptic NUP214::ABL1 Gene Fusion. Pathol. Oncol. Res. 28:1610570. doi: 10.3389/pore.2022.1610570
Received: 04 May 2022; Accepted: 02 September 2022;
Published: 12 September 2022.
Edited by:
Edit Bardi, St. Anna Kinderspital, AustriaCopyright © 2022 Stukaite-Ruibiene, Norvilas, Dirse, Stankeviciene and Vaitkeviciene. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Egle Stukaite-Ruibiene, ZWdsZS5lZ2xhaXRlQGdtYWlsLmNvbQ==
 Egle Stukaite-Ruibiene
Egle Stukaite-Ruibiene Rimvydas Norvilas2,3
Rimvydas Norvilas2,3