Abstract
Background: Human papillomavirus type 8 (HPV8) has been implicated in the progress of non-melanoma skin cancers and their precursor lesions. The HPV8 E7 oncoprotein plays a key role in the tumorigenesis of HPV-associated cutaneous tumors. However, the exact role of HPV8 E7 in human epidermal carcinogenesis has not been fully elucidated.
Methods: To investigate the potential carcinogenic effects of HPV8 E7 on epithelial cells, we used RNA-sequencing technology to analyze the gene expression profile of HPV8 E7-overexpressed normal human epidermal keratinocytes (NHEKs).
Results: RNA-sequencing revealed 831 differentially expressed genes (DEGs) between HPV8 E7-expressing NHEKs and control cells, among which, 631 genes were significantly upregulated, and 200 were downregulated. Gene ontology annotation enrichment analysis showed that HPV8 E7 mainly affected the expression of genes associated with protein heterodimerization activity, DNA binding, nucleosomes, and nucleosome assembly. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis revealed that overexpression of HPV8 E7 affected the expression of gene clusters associated with viral carcinogenesis and transcriptional misregulation in cancer and necroptosis signaling pathways that reportedly play crucial roles in HPV infection promotion and cancer progression. We also found the DEGs, such as HKDC1 and TNFAIP3, were associated with epigenetic modifications, immune regulation, and metabolic pathways.
Conclusion: Our results demonstrate that the pro-carcinogenic effect of HPV8 expression in epithelial cells may be attributed to the regulatory effect of oncogene E7 on gene expression associated with epigenetic modifications and immune and metabolic status-associated gene expression. Although our data are based on an in vitro experiment, it provides the theoretical evidence that the development of squamous cell carcinoma can be caused by HPV.
Introduction
Skin cancer incidence has rapidly increased over the years and is generally divided into malignant melanoma and non-melanoma skin cancer (NMSC), the latter being subdivided into cutaneous squamous cell carcinoma (SCC) and basal cell carcinoma [1]. To date, more than 200 human papillomavirus (HPV) genotypes have been discovered; these viruses can infect the skin and mucosa of several animal species [2]. HPVs are ubiquitous DNA viruses, and HPV infection, especially with high-risk subtypes, can mediate skin tumorigenesis [3]. HPV8, belonging to beta-genus HPV, is suspected to be carcinogenic in some skin cancer types, but the underlying pathological mechanisms of these cancers remain poorly understood [4].
HPV8 E6 and E7 reportedly promote the autophagic degradation of cellular checkpoint kinase-1, which may contribute to the oncogenic potential of the virus [5]. Furthermore, HPV8 E7 expression in the murine epidermis under the control of keratin-14 promoter showed that the E7 protein acts carcinogenically in mice by inducing the invasion of basal keratinocytes, which are regulated by the extracellular protein fibronectin [6]. However, the potential molecular mechanisms and the downstream transcriptional modulation effects of HPV8 E7-infected keratinocytes remain unclear [7, 8].
Changes in the epigenetic modifications, immune response, and metabolic status are associated with tumor occurrence due to HPV infection [9]. For example, clear CpG island methylation occurs in HPV-induced cutaneous warts [10]. Furthermore, recent studies have suggested that the HPV oncogenes E5, E6, and E7 play critical roles in the development of chronic inflammation through various mechanisms and can affect the redox homeostasis of host cells, inducing oxidative stress, which may promote viral integration and cancer development [11, 12]. Moreover, the oncoproteins HPV16 E6 and E7 regulate the cell metabolism to satisfy the energy demands necessary for persistent infection and development of cervical cancer [13]. However, the gene expression profiles of the oncogene HPV8 E7 in human epidermal keratinocytes remain unclear.
Currently, RNA-sequencing technology is widely used to detect transcriptome profiling in cancer research and therapy such as biomarker discovery and cancer heterogeneity, drug resistance, cancer microenvironment, and immunotherapy [14]. However, the transcriptome signatures in HPV8 E7-infected epithelial cells remain unclear. In our study, we took advantage of RNA-sequencing technology and the enrichment analysis of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) and discovered numerous significantly altered gene transcriptional landscapes that could be clustered into epigenetic modifications, immune regulations, and metabolic reprogramming. This study will elucidate the understanding of the tumorigenic mechanisms of HPV8 E7 expression.
Materials and Methods
Cells and Reagents
Normal human epidermal keratinocyte (NHEK) cells were purchased from the American Type Culture Collection (ATCC, No. PCS-200-010). EpiLife Media (60 µM calcium) supplemented with the HKGS kit (S001K, Thermo Fisher) was used for cell culture, and cells were cultured at 37°C in a 5% CO2 atmosphere. Complementary DNA (cDNA) for HPV8 E7 was amplified from the plasmid pCDNA3.1-HPV-8E7, which was a gift from Prof. Angel Alonso (German Cancer Research Centre, Heidelberg, Germany) and inserted into the P23 pHAGE-fEF1a-IRES-ZsGreen lentiviral vector with a C-terminal FLAG tag for lentiviral packaging.
Construction of Cell Line stably Overexpressing HPV8 E7
The HPV8 E7 overexpression cell line was constructed as described previously with a minor modification [15]. In brief, the HPV8 E7 lentivirus-overexpressing plasmid and packaging plasmids were co-transfected into HEK293T cells using the Lipofectamine 3000 transfection reagent for packaging. The supernatant of the cells was collected, combined, and centrifuged at 2,000 rpm for 5 min and then filtered using a filter with a diameter of 45 μm. The filtrate was determined by titer and stored at −80 °C until use. For lentiviral infection, 1 × 105 NHEK cells per well (a total of three independent wells) were seeded onto a 12-well plate overnight, and 1 ml of the lentiviral fluid was added to the cells for 4 h and 100 μg/ml of polybrene was added to enhance infection efficiency. For the control group, 1 × 105 NHEK cells per well (a total of three independent wells) were added with 1 ml of P23 pHAGE-fEF1a-IRES-ZsGreen control lentivirus. After 4 h, the virus medium was removed, and 1 ml of fresh complete medium was added. The infection efficiency was detected by flow cytometry after 48 h. The construct was confirmed via DNA sequencing.
RNA Extraction
TRIzol reagent (Invitrogen, Life Technologies, United States) was used to isolate the total RNAs from the NHEK cells infected with the HPV8 E7-overexpression lentivirus or the control virus, and the quality and concentration of the purified total RNAs were evaluated using a Nanodrop spectrophotometer (Thermo Scientific Technology, United States).
Library Construction and Sequencing
cDNA library construction and transcriptome sequencing were conducted by Lianchuan Biotechnology Co., Ltd. (Hangzhou, China) as previously described [16]. The Agilent Bioanalyzer 2100 (Santa Clara, CA, United States) was used to detect the RNA integrity. According to the standard procedure of Illumina (protocol # 15008136), 1 μg of the total RNA from each sample was used for each sequencing library. To minimize the potential ribosome RNA (rRNA) contamination, rRNAs were removed via a selection procedure using Poly (A) containing mRNA. The Experion DNA 1K chip was used to evaluate the libraries’ quality. After standard procedures including cDNA generation, end repair, A-tailing, adaptor ligation, and PCR amplification, the cDNAs were sequenced on an Illumina NovaSeq 6000 platform.
Functional and Pathway Enrichment Analysis of Differentially Expressed Genes
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) database was used for KEGG pathway and GO analyses of DEGs, as described previously [15, 17]. In brief, a false discovery rate with a threshold ≤0.05 was used to determine the p-value in multiple tests and ClusterProfiler R (3.4.4) software was used to visualize the KEGG pathway enrichment (http://www.genome.jp/kegg/) and GO terms (http://www.geneontology.org/).
Statistical Analysis
All statistical analyses were performed with GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, United States). Data were expressed as mean ± standard error of mean (S.E.M) and are representative of three independent experiments. The threshold criteria of DEGs were set as log2 fold-change value >1 and p-value < 0.05, and log (fold change) value >2. Significant differences were calculated by using an unpaired two-tailed Student’s t test, and p value < 0.05 was considered statistically significant.
Results
Overexpression of HPV8 E7 Affects the Gene Expression Profile in NHEKs
To investigate the potential carcinogenic effects of HPV8 E7 expression, we took advantage of RNA-sequencing technology to detect the gene expression profile of NHEK cells after HPV8 E7 overexpression and discovered 831 DEGs in the HPV8 E7-overexpressing NHEK cells compared with those in the controls (Figure 1A). Among the DEGs, 631 genes were significantly upregulated, and 200 were downregulated (Figure 1A). Furthermore, genes such as EGR1, PPP1R15A, JUN, and H2AC18 were found to be the most significantly upregulated genes and genes including AL109628, TRIM56, DLG3, and ATP6V1G2-DDX39B were the most significantly downregulated upon HPV8 E7 overexpression (Figures 1B,C). Fragments per kilobase of exon model per million mapped fragments (FPKM) values are shown in Figure 1D,E.
FIGURE 1
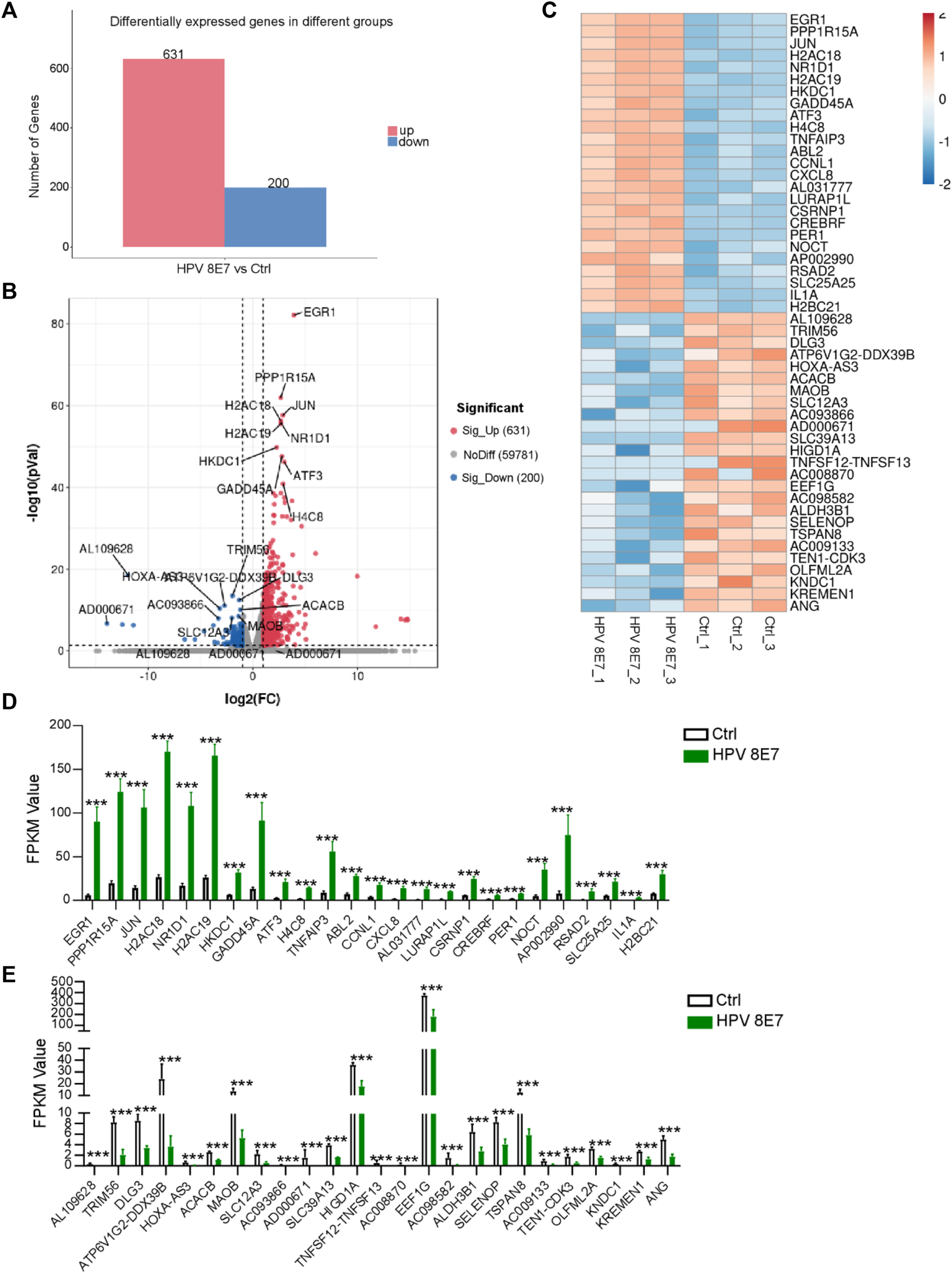
Overexpression of HPV8 E7 affected the gene expression profile in NHEKs. HPV8 E7 was overexpressed in NHEKs using the lentivirus system and the gene expression profiles were detected by the RNA-seq method. (A) Histogram representation of DEGs upregulated or downregulated in the control and HPV8 E7-overexpressed NHEK cells. (B,C) Volcano plot (B) and heatmap (C) representation of the 25 most significantly upregulated or downregulated genes in HPV8 E7-overexpressed NHEK cells compared with those in the controls. (D) Fragments per kilobase million (FPKM) values of the 25 most significantly upregulated genes in HPV8 E7-overexpressed NHEK cells compared with controls. (E) FPKM values of the 25 most significantly downregulated genes in HPV8 E7-overexpressed NHEK cells compared with those in the controls.
GO Enrichment Analysis of DEGs Affected by HPV8 E7 Overexpression
To determine the biological function of HPV8 E7 in NHEKs, GO analysis was performed to functionally annotate and classify the DEGs. According to the enrichment factor classification, protein heterodimerization activity, DNA binding, nucleosomes, and nucleosome assembly were the most significantly enriched (Figure 2A). All 44 DEGs, including A2AC18, H2AC19, GADD45, ATF3, and H4C8, associated with protein heterodimerization activity were dramatically increased after HPV8 E7 expression (Supplementary Figure S1A; Figure 2B). Furthermore, a total of 50 DEGs associated with DNA binding were detected, 39 among which were significantly upregulated, including EGR1, JUN, H2AC18, TNFAIP3, and CREB5; 11 genes were downregulated, including ZNF664, MYCN, PITX3, APOC0944, and WBP2NL in HPV8 E7-infected NHEKs compared with those in the controls (Supplementary Figure S1B; Figure 2C). HPV8 E7 inhibited 36 genes, including H2AC18, H2AC19, H4C8, H2BC21, and H2BC8, which could be enriched into nucleosomes (Supplementary Figure S1C; Figure 2D). Furthermore, 26 genes, including H4C8, H2BC21, H2BG8, H3C14, and H2BC4, associated with nucleosome assembly were markedly downregulated upon HPV8 E7 expression (Supplementary Figure S1D; Figure 2E).
FIGURE 2
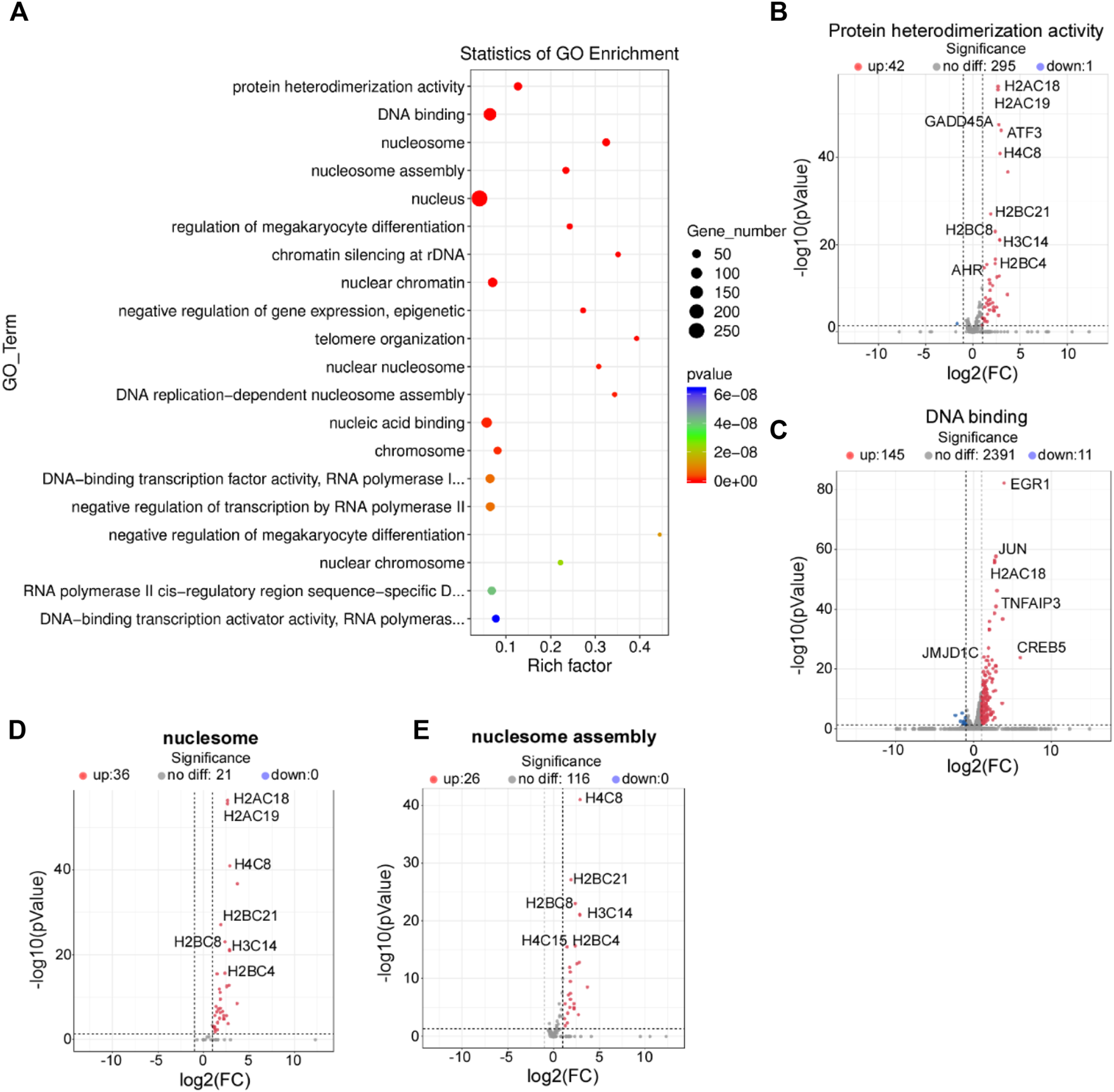
GO enrichment analysis of DEGs affected by HPV8 E7 overexpression. (A) Scatter plot for GO analysis. (B) Volcano plot representation of DEGs related to protein heterodimerization activity. (C) Volcano plot representation of DEGs related to DNA binding. (D) Volcano plot representation of DEGs related to nucleosomes. (E) Volcano plot representation of DEGs related to nucleosome assembly. See also Supplementary Figure S1.
KEGG Pathway Enrichment Analysis of DEGs Affected by HPV8 E7 Overexpression
To further investigate the epidermal pathways affected by HPV8 E7 expression, KEGG pathway enrichment analysis was conducted and revealed that HPV8 E7 overexpression mainly affected gene expression in the viral carcinogenesis pathway, transcriptional misregulation in cancer, and necroptosis pathways (Figure 3A). HPV8 E7 overexpression significantly inhibited SCIN expression but dramatically promoted the expression of 27 viral carcinogenesis-associated genes, including JUN, H4C8, H2C21, CREB5, and H2BC8, in NHEK cells (Supplementary Figure S2A; Figures 3B,C). On the other hand, in transcriptional misregulation in the cancer signaling pathway, only MYCN expression was notably decreased, and the expression of 20 genes, including GADD45A, CXCL8, JMJD1C, RUNX1, and H3C14, was significantly increased in NHEKs upon HPV8 E7 expression (Supplementary Figure S2B; Figures 3D,E). Further, HPV8 E7 overexpression significantly inhibited TICAM2 expression but substantially promoted the expression of 15 genes, including H2AC18, H2AC19, TNFAIP3, IL1A, and CYLD in NHEK cells, all of which were crucial for the necroptosis signaling pathway (Supplementary Figure S2C; Figures 3F,G).
FIGURE 3
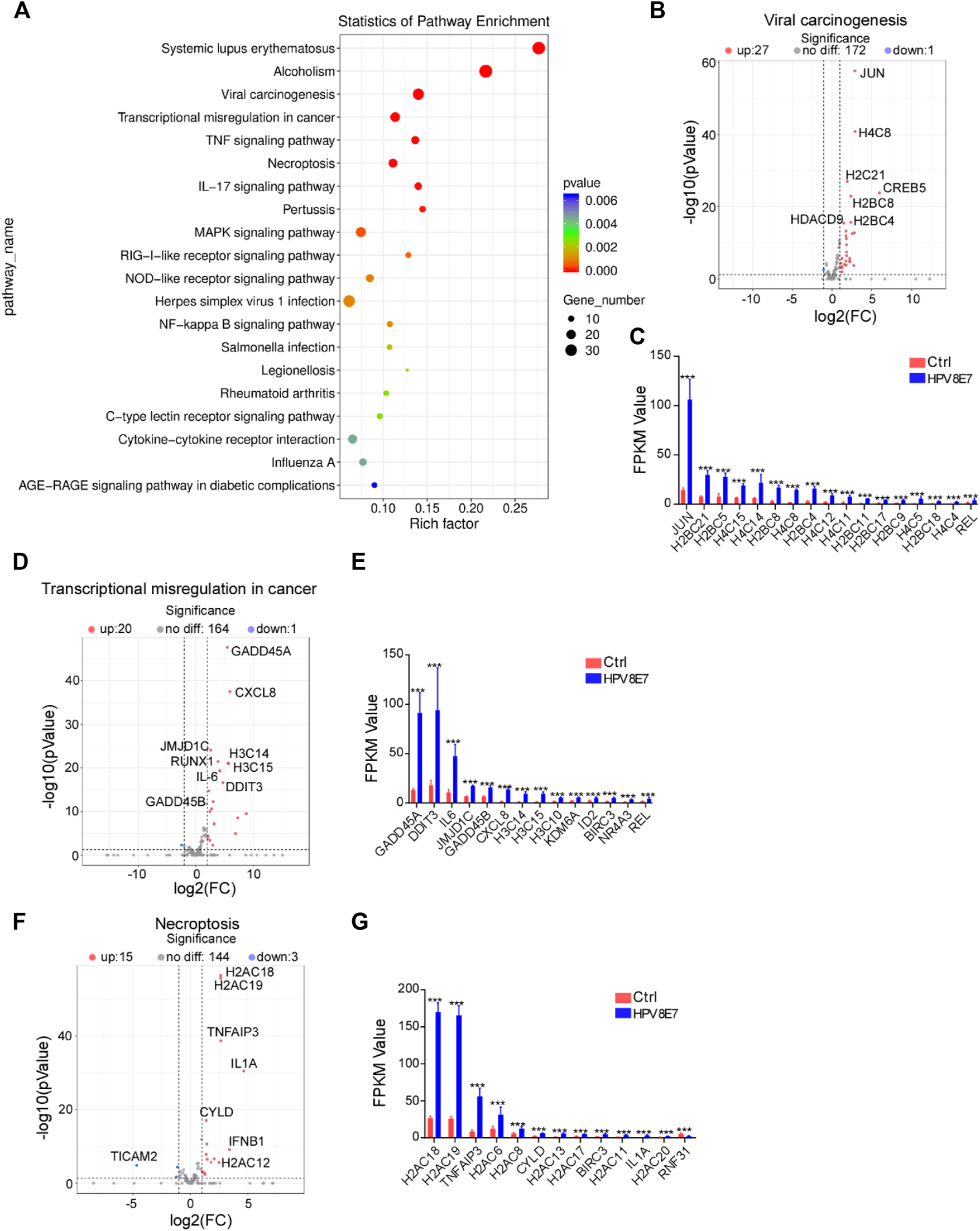
KEGG pathway enrichment analysis of DEGs affected by HPV8 E7 overexpression. (A) Scatter plot showing the enrichment analysis of the KEGG pathway. (B,C) Volcano plot (B) and FPKM value >1 (B) representation of DEGs related to viral carcinogenesis. (D,E) Volcano plot (D) and FPKM value >1 (E) representation of DEGs related to transcriptional misregulation in cancer. (F,G) Volcano plot (F) and FPKM value >1 (G) representation of DEGs related to necroptosis. See also Supplementary Figure S2.
HPV8 E7 Overexpression Significantly Affected Metabolism-Associated Gene Expression
To evaluate the regulatory effects of HPV8 E7 on the metabolism status in epithelial cells, the cell metabolism-associated genes were enriched in NHEK cells after HPV8 E7 expression. As shown in Figures 4A,B, 23 DEGs associated with the metabolism pathway were detected in HPV8 E7-overexpressed NHEKs compared with controls, 11 of which were notably downregulated, such as PLA2G4B, AK4, OSBPL5, ACACB, and AK7; 12 genes were dramatically upregulated, including JUN, HKDC1, PIP5K1A, and MGAM. HPV8 E7 overexpression significantly inhibited PLASG4B expression but promoted PISD, ADPRM, PLA2G4C, and ACHE expression, which was associated with the glycerophospholipid metabolism pathway (Supplementary Figure S3A; Figures 4C,D). Furthermore, HPV8 E7 markedly decreased ALDH3B1 expression, but increased HKDC1 and ENO3 expression, which was associated with the glycolysis metabolism pathway (Supplementary Figure S3B; Figures 4E,F). Moreover, HPV8 E7 overexpression significantly inhibited the expression of PIP5K1A, IPMK, and PLCB4, which was associated with the inositol phosphate metabolism pathway (Supplementary Figure S3C; Figures 4G,H). These results indicated that HPV8 E7 expression significantly altered the metabolism status in epidermal keratinocytes.
FIGURE 4
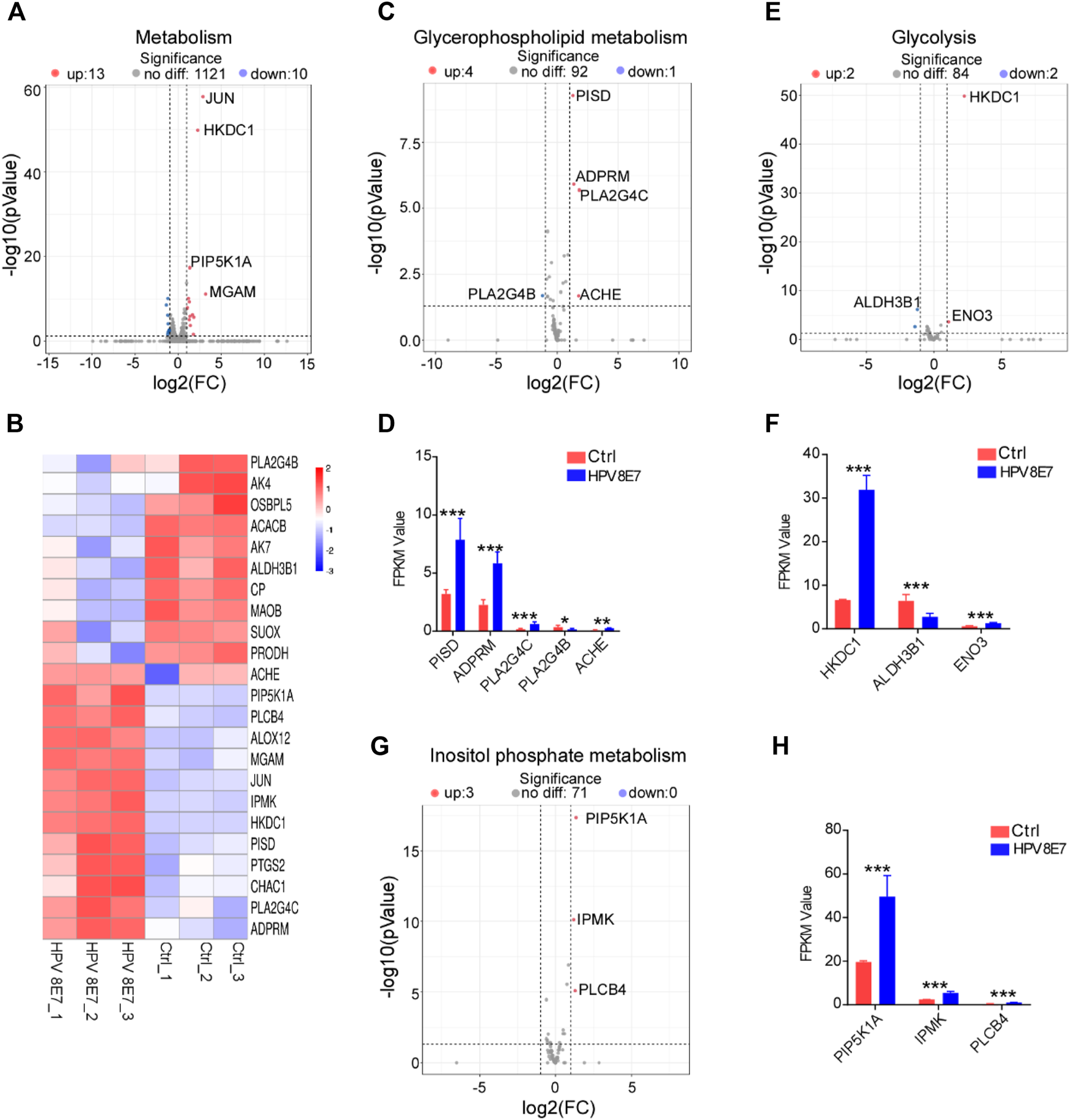
HPV8 E7 overexpression significantly affected metabolism-associated gene expression. (A,B) Volcano plot (A) and heatmap (B) representation of DEGs related to metabolism. (C,D) Volcano plot (C) and FPKM value > 1 (D) representation of DEGs related to glycerophospholipid metabolism. (E,F) Volcano plot (E) and FPKM value > 1 (F) representation of DEGs related to glycolysis. (G,H) Volcano plot (G) and FPKM value (H) representation of DEGs related to inositol phosphate metabolism. See also Supplementary Figure S3.
HPV8 E7 Overexpression Significantly Affected Epigenetic-Associated Gene Expression
To evaluate the regulatory effects of HPV8 E7 on epigenetic modifications in epithelial cells, cell epigenetic-associated genes were enriched in NHEK cells after HPV8 E7 expression. As shown in Supplementary Figure S4A; Figures 5A,B, DNA and RNA methylation-associated genes, such as NSUN6, RLF, RBM15, and PIWIL2 were dramatically upregulated after HPV8 E7 expression. Furthermore, HPV8 E7 overexpression significantly promoted the expression of histone acetylation and deacetylation-associated genes, such as SNAI2, FLCN, and HDAC9 (Supplementary Figure S4B; Figures 5C,D). Moreover, HPV8 E7 expression also notably increased the expression of the histone methylation and demethylation-associated genes, including JMJD1C, KDM6A, and KDM6B (Supplementary Figure S4C; Figures 5E,F). These results indicated that HPV8 E7 expression significantly increased the overall epigenetic modifications of epithelial cells.
FIGURE 5
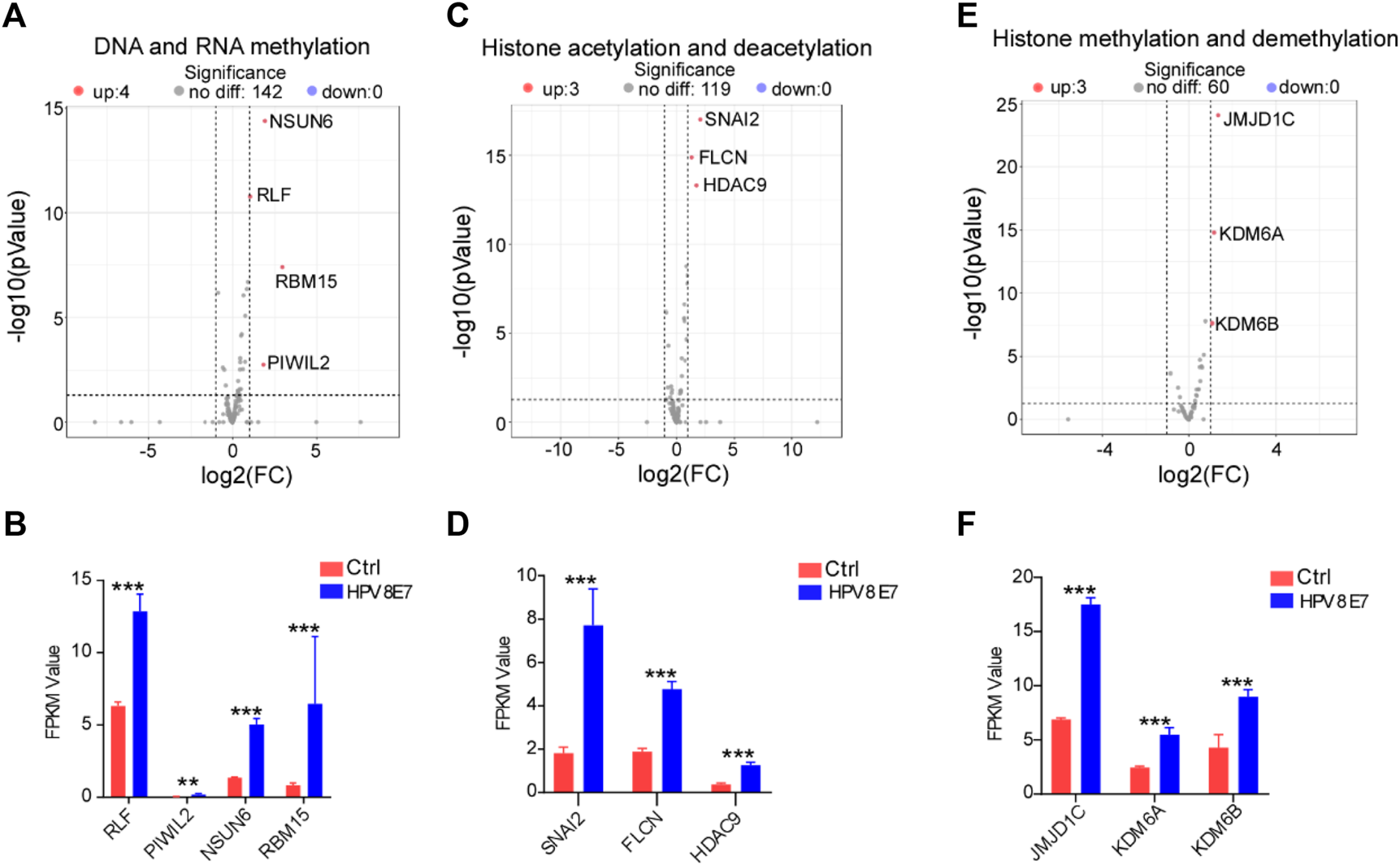
HPV8 E7 overexpression significantly affected epigenetic-associated gene expression. (A,B) Volcano plot (A) and FPKM value (B) representation of DEGs related to DNA and RNA methylation. (C,D) Volcano plot (C) and FPKM value (D) representation of DEGs related to histone acetylation and deacetylation. (E,F) Volcano plot (E) and FPKM value (F) representation of DEGs related to histone methylation and demethylation. See also Supplementary Figure S4.
HPV8 E7 Overexpression Significantly Affected Immune Response-Associated Gene Expression
Inflammatory response plays a pivotal role in the defense against viral infection. To evaluate the immune status after HPV8 E7 expression, immune response signaling pathway-associated genes were enriched. As shown in Figure 6A, 66 DEGs were detected in the NHEK cells after HPV8 E7 expression, among which, the expression of 8 genes, such as TICAM2, TRIM56, C5, AQP3, and CACNA1C, was significantly inhibited, but that of 58 genes, including TNFAIP3, CXCL8, RSAD2, IL1A, and H2BC21 was substantially increased (Figures 6B,C). These results indicated that HPV8 E7 expression may affect an intense immune response in epithelial keratinocytes.
FIGURE 6
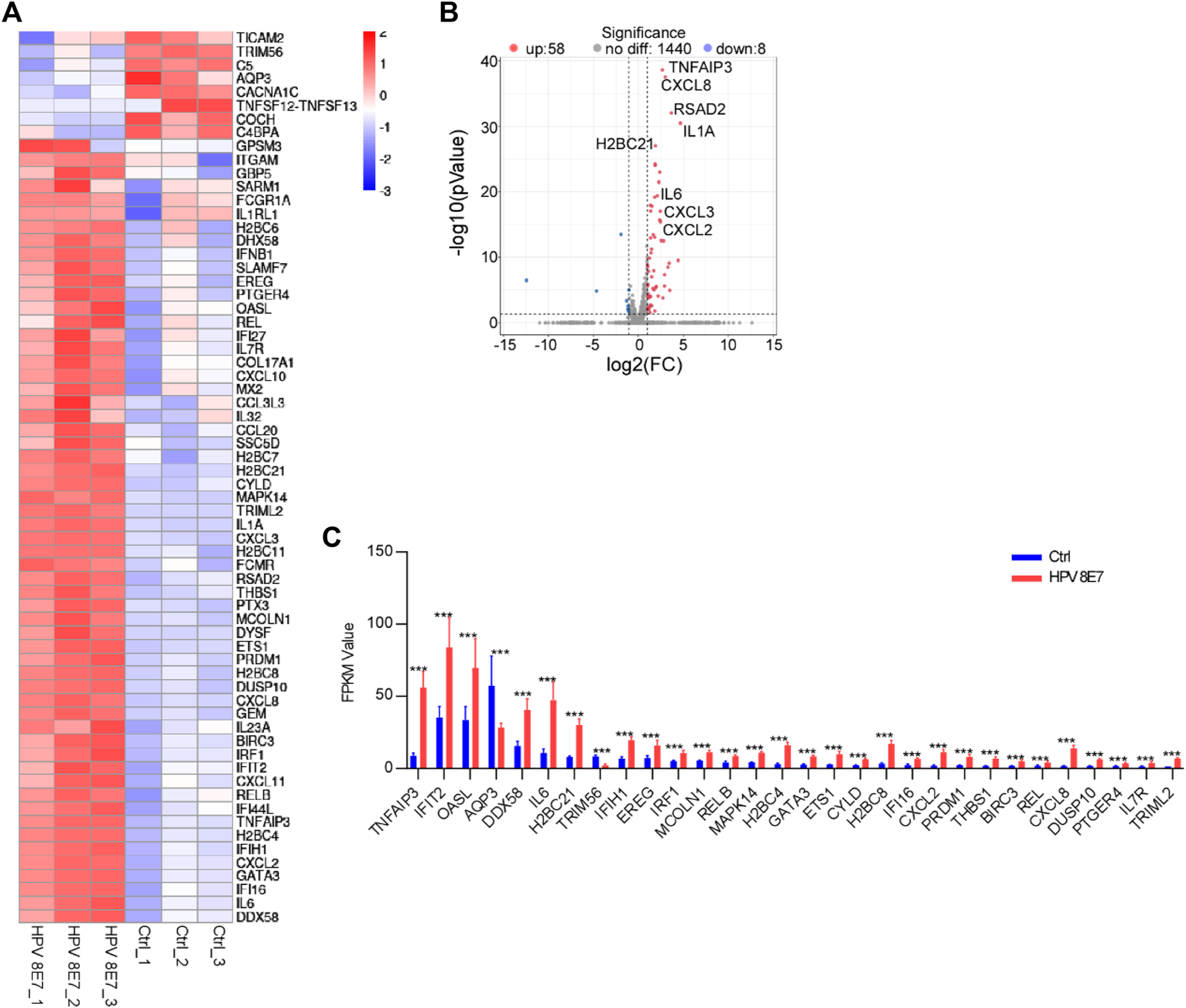
HPV8 E7 overexpression significantly affected immune response-associated gene expression. (A) Heatmap representation of the DEGs related to immunoregulation. (B) Volcano plot representation of the DEGs related to immunoregulation. (C) FPKM value (>1) of the DEGs related to immunoregulation.
Discussion
HPV infection, especially with high-risk subtypes, is associated with SCCs that arise in several different organs but share many common biological and immunological properties [18]. In cervical and anal cancers, HPV is the etiology for nearly 90% of malignancies, whereas only 30% of oropharyngeal, vaginal, vulvar, and penile cancers are driven by carcinogen exposure or other causes [19]. Mechanistically, HPV oncogenes not only directly inhibit the tumor suppressors p53 and Rb, but also suppress apoptosis and telomere shortening and promote genomic instability, angiogenesis, invasion, and metastasis to facilitate malignant transformation [20–22]. These properties render HPV infection, induced by factors other than sunlight exposure/UV light, or chemical irritants, an ideal model for the study of the potential mechanisms of HPV-associated cancers.
The functional evaluation of HPVs is far from complete, but currently, the two viral oncoproteins E6 and E7 have been the focus [23]. The HPV8 E6 oncoprotein mainly impairs the DNA damage repair pathway to facilitate carcinogenesis [24]. Moreover, carcinogenic effects are observed in epidermis HPV8 E7 oncoprotein-overexpressing transgenic mice [6]. In vitro, HPV8 E6 counters the cellular response to DNA damage and mitotic errors by destabilizing the functions of P300, a histone acetyltransferase [25]. Furthermore, a recent study finds that HPV8 E7-expressing human primary keratinocytes increase GADD34 and GDF15 expression to facilitate skin tumorigenesis by activating the Src kinase family members Fyn and Lyn [26]. Unfortunately, although the HPV8 E7 oncogene has been identified in more than 90% of SCCs and has been implicated in NMSCs, its exact role in human skin carcinogenesis has not been fully elucidated [3, 27, 28]. In this study, we took advantage of RNA-sequencing technology and the enrichment analysis of GO and KEGG pathways and found that 831 genes were significantly altered upon HPV8 E7 overexpression. Most of the DEGs could be clustered into nucleosomes, nucleosome assembly, viral carcinogenesis pathway, transcriptional misregulation in cancer, and necroptosis pathways. These are the first in vitro mechanistic data on the tumorigenicity of HPV8 E7 and may shed new light on the tumorigenic effects of HPV8 E7 expression.
The GO resource (http://geneontology.org) is a structured, computable database on gene function [29]. In the present study, most HPV8 E7 overexpression-induced genes could be clustered into protein heterodimerization activity and DNA binding (molecular function), nucleosomes (cellular component), and nucleosome assembly (biological process). Our results support the hypothesis that viral proteins interact and manipulate not only cytoplasmic components but also nuclear factors for their survival and propagation [30, 31]. The KEGG pathway database is a reference knowledge base that helps reconstruct molecular network systems [32]. In this study, the KEGG pathway enrichment analysis revealed that HPV8 E7 overexpression mainly affected gene expression in the viral carcinogenesis pathway, transcriptional misregulation in cancer, and necroptosis pathways, which were either important for HPV infection-driven carcinogenesis or could be an important marker for diagnosis and prognosis in HPV-related cancer [33, 34].
Metabolic reprogramming is an important hallmark in cancer [13]. However, the mechanisms of how HPV viral oncoproteins alter cell metabolism to satisfy their own energy demands and how this may contribute to tumorigenesis remain unknown [35]. Interestingly, the spare mitochondrial respiratory capacity of HPV8 E7-expressing cells is markedly increased, and this effect may be due to the direct interaction between HPV8 E7 and the mitochondrial ATP-synthase, which would dramatically result in decreased glycolytic activity [35]. Furthermore, the function of cell metabolism biomolecules, such as glucose, amino acids, and lipids, has been altered to promote cell proliferation, inhibit cell death, and affect genomic instability after HPV infection [9]. Indeed, in the current study, we not only detected DEG profiles in glycolysis pathways in HPV8 E7-expressing NHEKs, but also clustered DEGs into glycerophospholipid metabolism and inositol phosphate metabolism signaling pathways. Glycerophospholipids, the major components of cell membranes, play an important role in viral infection and are crucial for the morphogenesis of progeny virus and viral pathogenicity [36, 37]. Moreover, the G/G genotype and G allele of the inositol 1,4,5-trisphosphate 3-kinase C rs28493229 polymorphism may raise the risk of development of cervical SCC, and could be a potential marker for genetic susceptibility to CSCC [38].
It has been reported viral proteins can regulate epigenetic modifications to affect biological functions, such as cell proliferation and apoptosis [39]. Epigenetic mechanisms related to HPV infection include methylation changes to host and viral DNA and chromatin modification in host species [40]. In this study, we detected 10 epigenetic-associated genes significantly upregulated upon HPV8 E7 overexpression in epidermal keratinocytes, and these DEGs were clustered into DNA and RNA methylation, histone acetylation and deacetylation, and histone methylation and demethylation. HPV E7 promotes the expression of SUV39H1, which is a chromatin repressor and can inhibit the mRNA levels of RIG-I, cGAS, and STING genes by epigenetic silencing, and thereby blocks the interferon secretion in HPV-transformed cells [41]. Furthermore, changed methylated CpGs and regions reportedly regulate gene expression and affect prognosis in HPV-positive head and neck SCC [42]. Further, the epigenetic regulators histone deacetylases (1 and 2) are overexpressed in oncoprotein E6/E7-expressing cells, and histone deacetylase inhibitors could be a therapeutic intervention for skin cancer induced by HPV [43, 44]. An epigenome-wide analysis of HPV-driven common warts reveals that H3F3A, CDKN1A, and MAPK13 are the most differentially methylated genes [45]. Furthermore, HPV viral proteins can directly target histone methyltransferases for activity inhibition and modulate specific gene expression [39].
While the association between global immune compromise and HPV-related malignancies is well established, our understanding of how HPV perpetuates a state of immune suppression in the microenvironment remains limited [18]. Viral oncoproteins can control various cellular circuit programs, such as innate immunity to promote the proliferation of HPV-transformed cells [41]. However, the mechanisms of how persistent HPV8 infection affects the immune response to promote the carcinogenic process remain unknown. HPV E7 promotes the expression of H3K9-specific methyltransferase, which can regulate the host innate immune response to facilitate immune evasion [32]. However, in this study, we found many acute immune response genes, such as TNFAIP3, and those encoding interleukins and chemokines expression in HPV8 E7-expressing NHEKs, which were associated with the clearance of HPV infection.
However, the present study only provided the transcriptional regulation landscape of HPV8 E7-expressing NHEKs; our study limitation is that data interpretation is based only on an in vitro experiment and that it provides only theoretical evidence. Further, these experiments were conducted on a single keratinocyte, and whether the same results can be obtained from normal keratinocytes of other species remains uncertain; therefore, the exact tumorigenic mechanisms of HPV8 E7 need to be further corroborated.
Conclusion
In conclusion, our results demonstrated the pro-carcinogenic effect of HPV8 expression in epithelial cells which could contribute to the regulatory effect of oncogene E7 on gene expression associated with nucleosomes, nucleosome assembly, viral carcinogenesis pathway, transcriptional misregulation in cancer, necroptosis pathways, epigenetic modifications, immune response, and metabolic status. Although our data are based on an in vitro experiment, it provides more effective evidence that the development of squamous cell carcinoma can be caused by HPV.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XC, YT, and QL conceived and designed the study. XC, ML, YT, and CH performed the experiments. XC, YT, and HH analyzed the data. XC and YS wrote and reviewed the manuscript. XC, YS, and HC have overall responsibility for this manuscript. YS and HC were responsible for supervision and funding acquisition.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81000698, 81573057, and 81703135), the Natural Science Foundation of Zhejiang Province (LQ19H030006), and the Medical and Health Science and Technology Project of Zhejiang Province (2018256428).
Acknowledgments
We thank the Core Facilities, Zhejiang University School of Medicine and the Key Laboratory of Immunity and Inflammatory Diseases of Zhejiang Province for technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2022.1610176/full#supplementary-material
References
1.
Mitsiogianni M Amery T Franco R Zoumpourlis V Pappa A Panayiotidis MI From Chemo-Prevention to Epigenetic Regulation: The Role of Isothiocyanates in Skin Cancer Prevention. Pharmacol Ther (2018) 190:187–201. 10.1016/j.pharmthera.2018.06.001
2.
Schiffman M Doorbar J Wentzensen N de Sanjosé S Fakhry C Monk BJ et al Carcinogenic Human Papillomavirus Infection. Nat Rev Dis Primers (2016) 2:16086. 10.1038/nrdp.2016.86
3.
Smola S. Human Papillomaviruses and Skin Cancer. Adv Exp Med Biol (2020) 1268:195–209. 10.1007/978-3-030-46227-7_10
4.
Ding X Lucas T Marcuzzi GP Pfister H Eming SA. Distinct Functions of Epidermal and Myeloid-Derived VEGF-A in Skin Tumorigenesis Mediated by HPV8. Cancer Res (2015) 75:330–43. 10.1158/0008-5472.CAN-13-3007
5.
Akgül B Kirschberg M Storey A Hufbauer M. Human Papillomavirus Type 8 Oncoproteins E6 and E7 Cooperate in Downregulation of the Cellular Checkpoint Kinase-1. Int J Cancer (2019) 145:797–806. 10.1002/ijc.32223
6.
Heuser S Hufbauer M Steiger J Marshall J Sterner-Kock A Mauch C et al The Fibronectin/α3β1 Integrin axis Serves as Molecular Basis for Keratinocyte Invasion Induced by βHPV. Oncogene (2016) 35:4529–39. 10.1038/onc.2015.512
7.
Tommasino M. The Biology of Beta Human Papillomaviruses. Virus Res (2017) 231:128–38. 10.1016/j.virusres.2016.11.013
8.
Pfister H. HPV und Neoplasien der Haut. Hautarzt (2008) 59:26–30. 10.1007/s00105-007-1442-6
9.
Cruz-Gregorio A Aranda-Rivera AK Ortega-Lozano AJ Pedraza-Chaverri J Mendoza-Hoffmann F. Lipid Metabolism and Oxidative Stress in HPV-Related Cancers. Free Radic Biol Med (2021) 172:226–36. 10.1016/j.freeradbiomed.2021.06.009
10.
Al-Eitan LN Alghamdi MA Tarkhan AH Al-Qarqaz FA. Genome-Wide CpG Island Methylation Profiles of Cutaneous Skin with and without HPV Infection. Int J Mol Sci (2019) 20:4822. 10.3390/ijms20194822
11.
Georgescu SR Mitran CI Mitran MI Caruntu C Sarbu MI Matei C et al New Insights in the Pathogenesis of HPV Infection and the Associated Carcinogenic Processes: The Role of Chronic Inflammation and Oxidative Stress. J Immunol Res (2018) 2018:1–10. 10.1155/2018/5315816
12.
Liu X Ma X Lei Z Feng H Wang S Cen X et al Chronic Inflammation-Related HPV: A Driving Force Speeds Oropharyngeal Carcinogenesis. PLoS One (2015) 10:e0133681. 10.1371/journal.pone.0133681
13.
Arizmendi-Izazaga A Navarro-Tito N Jiménez-Wences H Mendoza-Catalán MA Martínez-Carrillo DN Zacapala-Gómez AE et al Metabolic Reprogramming in Cancer: Role of HPV 16 Variants. Pathogens (2021) 10:347. 10.3390/pathogens10030347
14.
Wang Y Mashock M Tong Z Mu X Chen H Zhou X et al Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front Oncol (2020) 10:447. 10.3389/fonc.2020.00447
15.
Zheng H Zou Z Wu X Xu Y Zhu J Zhou Q et al HPV11E7 Inhibits IMQ-Induced Chemokine and colony-stimulating Factor Production in Keratinocytes. Gene (2020) 760:145003. 10.1016/j.gene.2020.145003
16.
Zhang J Zhang C Sun P Huang M Fan M Liu M. RNA-sequencing and Pathway Analysis Reveal Alteration of Hepatic Steroid Biosynthesis and Retinol Metabolism by Tributyltin Exposure in Male Rare Minnow (Gobiocypris Rarus). Aquat Toxicol (2017) 188:109–18. 10.1016/j.aquatox.2017.03.015
17.
Wang T Zhang Y Guo Y Zhang X Yang H Tian X et al RNA-sequence Reveals Differentially Expressed Genes Affecting the Crested Trait of Wumeng Crested Chicken. Poult Sci (2021) 100:101357. 10.1016/j.psj.2021.101357
18.
Shamseddine AA Burman B Lee NY Zamarin D Riaz N. Tumor Immunity and Immunotherapy for HPV-Related Cancers. Cancer Discov (2021) 11:1896–912. 10.1158/2159-8290.CD-20-1760
19.
Saraiya M Unger ER Thompson TD Lynch CF Hernandez BY Lyu CW et al US Assessment of HPV Types in Cancers: Implications for Current and 9-valent HPV Vaccines. J Natl Cancer Inst (2015) 107:djv086. 10.1093/jnci/djv086
20.
Roden RBS Stern PL. Opportunities and Challenges for Human Papillomavirus Vaccination in Cancer. Nat Rev Cancer (2018) 18:240–54. 10.1038/nrc.2018.13
21.
Pal A Kundu R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front Microbiol (2019) 10:3116. 10.3389/fmicb.2019.03116
22.
McLaughlin-Drubin ME Münger K. The Human Papillomavirus E7 Oncoprotein. Virology (2009) 384:335–44. 10.1016/j.virol.2008.10.006
23.
Kyrgiou M Moscicki A-B. Vaginal Microbiome and Cervical Cancer. Semin Cancer Biol (2022). 10.1016/j.semcancer.2022.03.005
24.
Hufbauer M Cooke J van der Horst GTJ Pfister H Storey A Akgül B. Human Papillomavirus Mediated Inhibition of DNA Damage Sensing and Repair Drives Skin Carcinogenesis. Mol Cancer (2015) 14:183. 10.1186/s12943-015-0453-7
25.
Wendel SO Wallace NA. Loss of Genome Fidelity: Beta HPVs and the DNA Damage Response. Front Microbiol (2017) 8:2250. 10.3389/fmicb.2017.02250
26.
Kirschberg M Syed AS Dönmez HG Heuser S Wilbrand-Hennes A Alonso A et al Novel Insights into Cellular Changes in HPV8-E7 Positive Keratinocytes: A Transcriptomic and Proteomic Analysis. Front Microbiol (2021) 12:672201. 10.3389/fmicb.2021.672201
27.
Haedicke J Iftner T. Human Papillomaviruses and Cancer. Radiother Oncol (2013) 108:397–402. 10.1016/j.radonc.2013.06.004
28.
Oswald E Reinz E Voit R Aubin F Alonso A Auvinen E. Human Papillomavirus Type 8 E7 Protein Binds Nuclear Myosin 1c and Downregulates the Expression of Pre-rRNA. Virus Genes (2017) 53:807–13. 10.1007/s11262-017-1491-6
29.
The Gene Ontology C. The Gene Ontology Resource: 20 Years and Still Going Strong. Nucleic Acids Res (2019) 47:D330–D338. 10.1093/nar/gky1055
30.
Sultana S Zarreen F Chakraborty S. Insights into the Roles of Histone Chaperones in Nucleosome Assembly and Disassembly in Virus Infection. Virus Res (2021) 297:198395. 10.1016/j.virusres.2021.198395
31.
Pismataro MC Felicetti T Bertagnin C Nizi MG Bonomini A Barreca ML et al 1,2,4-Triazolo[1,5-a]pyrimidines: Efficient One-step Synthesis and Functionalization as Influenza Polymerase PA-PB1 Interaction Disruptors. Eur J Med Chem (2021) 221:113494. 10.1016/j.ejmech.2021.113494
32.
Kanehisa M Furumichi M Sato Y Ishiguro-Watanabe M Tanabe M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res (2021) 49:D545–D551. 10.1093/nar/gkaa970
33.
Akgül B Ghali L Davies D Pfister H Leigh IM Storey A. HPV8 Early Genes Modulate Differentiation and Cell Cycle of Primary Human Adult Keratinocytes. Exp Dermatol (2007) 16:590–9. 10.1111/j.1600-0625.2007.00569.x
34.
Li L Yu S Zang C. Low Necroptosis Process Predicts Poor Treatment Outcome of Human Papillomavirus Positive Cervical Cancers by Decreasing Tumor-Associated Macrophages M1 Polarization. Gynecol Obstet Invest (2018) 83:259–67. 10.1159/000487434
35.
Kirschberg M Heuser S Marcuzzi GP Hufbauer M Seeger JM Đukić A et al ATP Synthase Modulation Leads to an Increase of Spare Respiratory Capacity in HPV Associated Cancers. Sci Rep (2020) 10:17339. 10.1038/s41598-020-74311-6
36.
Arii J Fukui A Shimanaka Y Kono N Arai H Maruzuru Y et al Role of Phosphatidylethanolamine Biosynthesis in Herpes Simplex Virus 1-Infected Cells in Progeny Virus Morphogenesis in the Cytoplasm and in Viral Pathogenicity In Vivo. J Virol (2020) 94:e01572. 10.1128/JVI.01572-20
37.
Guardado-Calvo P Atkovska K Jeffers SA Grau N Backovic M Pérez-Vargas J et al A Glycerophospholipid-specific Pocket in the RVFV Class II Fusion Protein Drives Target Membrane Insertion. Science (2017) 358:663–7. 10.1126/science.aal2712
38.
Yang Y-C Chang T-Y Chen T-C Chang S-C Chen W-F Chan H-W et al Genetic Polymorphisms in the ITPKC Gene and Cervical Squamous Cell Carcinoma Risk. Cancer Immunol Immunother (2012) 61:2153–9. 10.1007/s00262-012-1280-y
39.
Hsu C-H Peng K-L Jhang H-C Lin C-H Wu S-Y Chiang C-M et al The HPV E6 Oncoprotein Targets Histone Methyltransferases for Modulating Specific Gene Transcription. Oncogene (2012) 31:2335–49. 10.1038/onc.2011.415
40.
Di Domenico M Giovane G Kouidhi S Iorio R Romano M De Francesco F et al HPV Epigenetic Mechanisms Related to Oropharyngeal and Cervix Cancers. Cancer Biol Ther (2018) 19:850–7. 10.1080/15384047.2017.1310349
41.
Lo Cigno I Calati F Borgogna C Zevini A Albertini S Martuscelli L et al Human Papillomavirus E7 Oncoprotein Subverts Host Innate Immunity via SUV39H1-Mediated Epigenetic Silencing of Immune Sensor Genes. J Virol (2020) 94:e01812–e01819. 10.1128/JVI.01812-19
42.
Degli Esposti D Sklias A Lima SC Beghelli-de la Forest Divonne S Cahais V Fernandez-Jimenez N et al Unique DNA Methylation Signature in HPV-Positive Head and Neck Squamous Cell Carcinomas. Genome Med (2017) 9:33. 10.1186/s13073-017-0419-z
43.
Roy M Mukherjee S. Reversal of Resistance towards Cisplatin by Curcumin in Cervical Cancer Cells. Asian Pac J Cancer Prev (2014) 15:1403–10. 10.7314/apjcp.2014.15.3.1403
44.
Lourenço de Freitas N Deberaldini MG Gomes D Pavan AR Sousa  Dos Santos JL et al Histone Deacetylase Inhibitors as Therapeutic Interventions on Cervical Cancer Induced by Human Papillomavirus. Front Cel Dev. Biol. (2020) 8:592868. 10.3389/fcell.2020.592868
45.
Al-Eitan LN Alghamdi MA Tarkhan AH Al-Qarqaz FA. Epigenome-wide Analysis of Common Warts Reveals Aberrant Promoter Methylation. Int J Med Sci (2020) 17:191–206. 10.7150/ijms.39261
Summary
Keywords
RNA-sequencing, HPV 8E7, immune regulations, metabolic reprogramming, epigenetic modifications
Citation
Chen X, Li M, Tang Y, Liang Q, Hua C, He H, Song Y and Cheng H (2022) Gene Expression Profile Analysis of Human Epidermal Keratinocytes Expressing Human Papillomavirus Type 8 E7. Pathol. Oncol. Res. 28:1610176. doi: 10.3389/pore.2022.1610176
Received
04 November 2021
Accepted
21 April 2022
Published
18 May 2022
Volume
28 - 2022
Edited by
Krenács Tibor, Semmelweis University, Hungary
Updates
Copyright
© 2022 Chen, Li, Tang, Liang, Hua, He, Song and Cheng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinjing Song, 3315023@zju.edu.cn; Hao Cheng, chenghao1@zju.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.